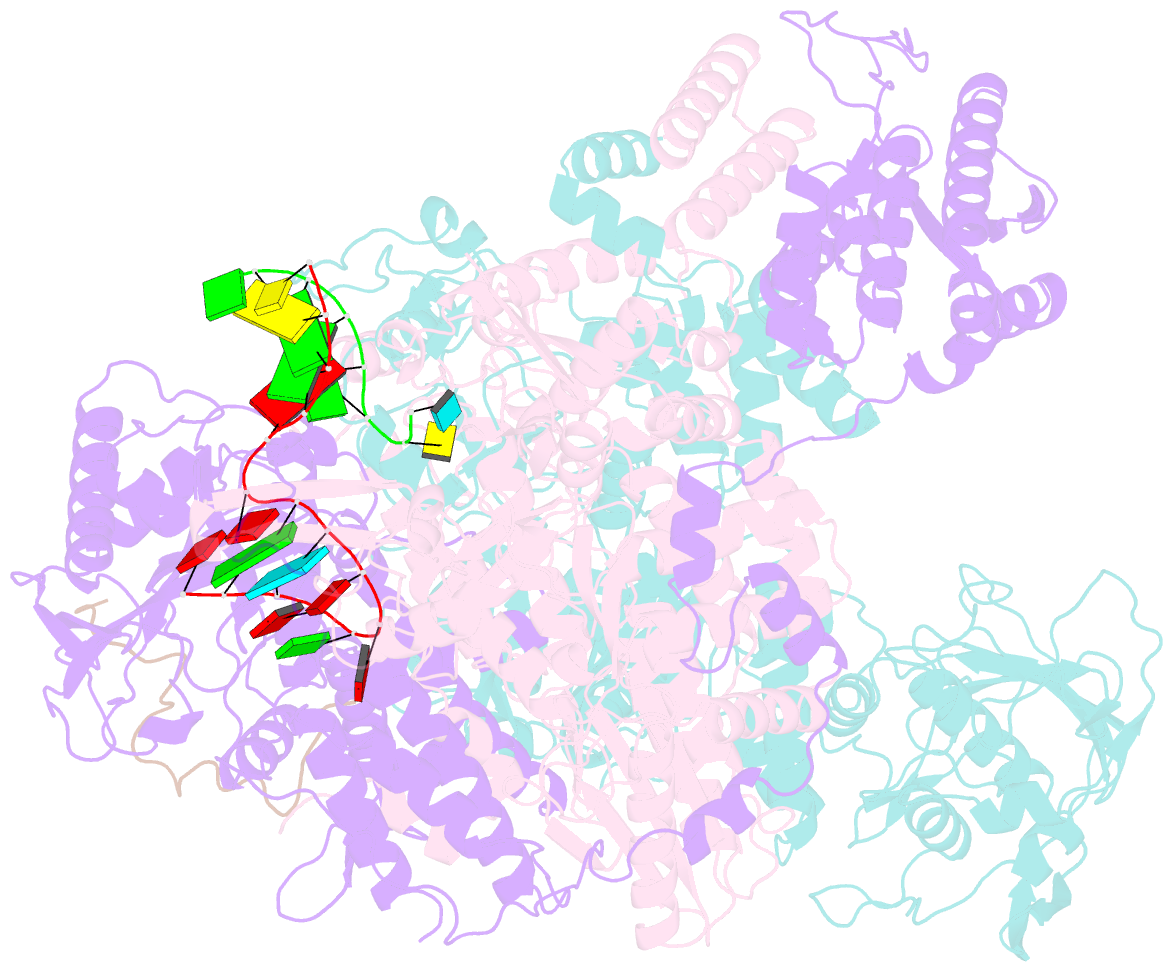
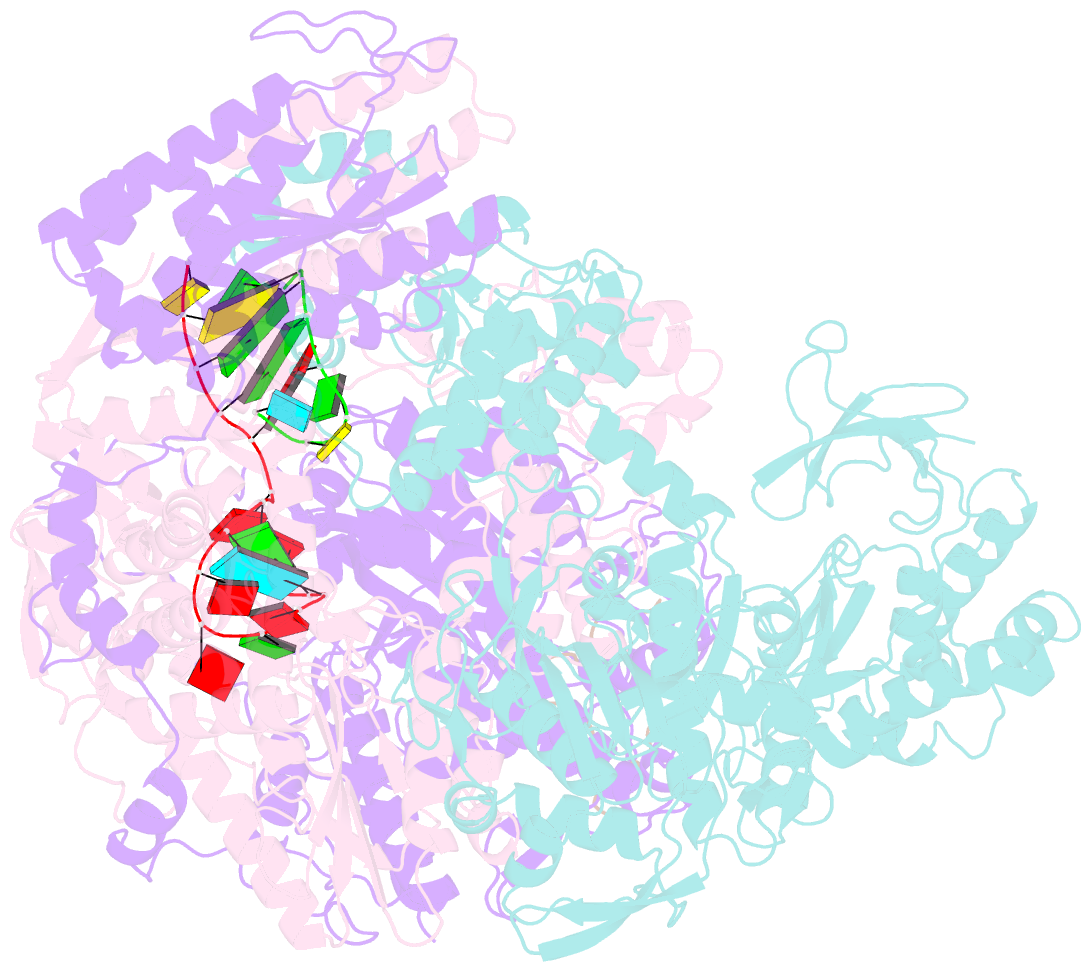
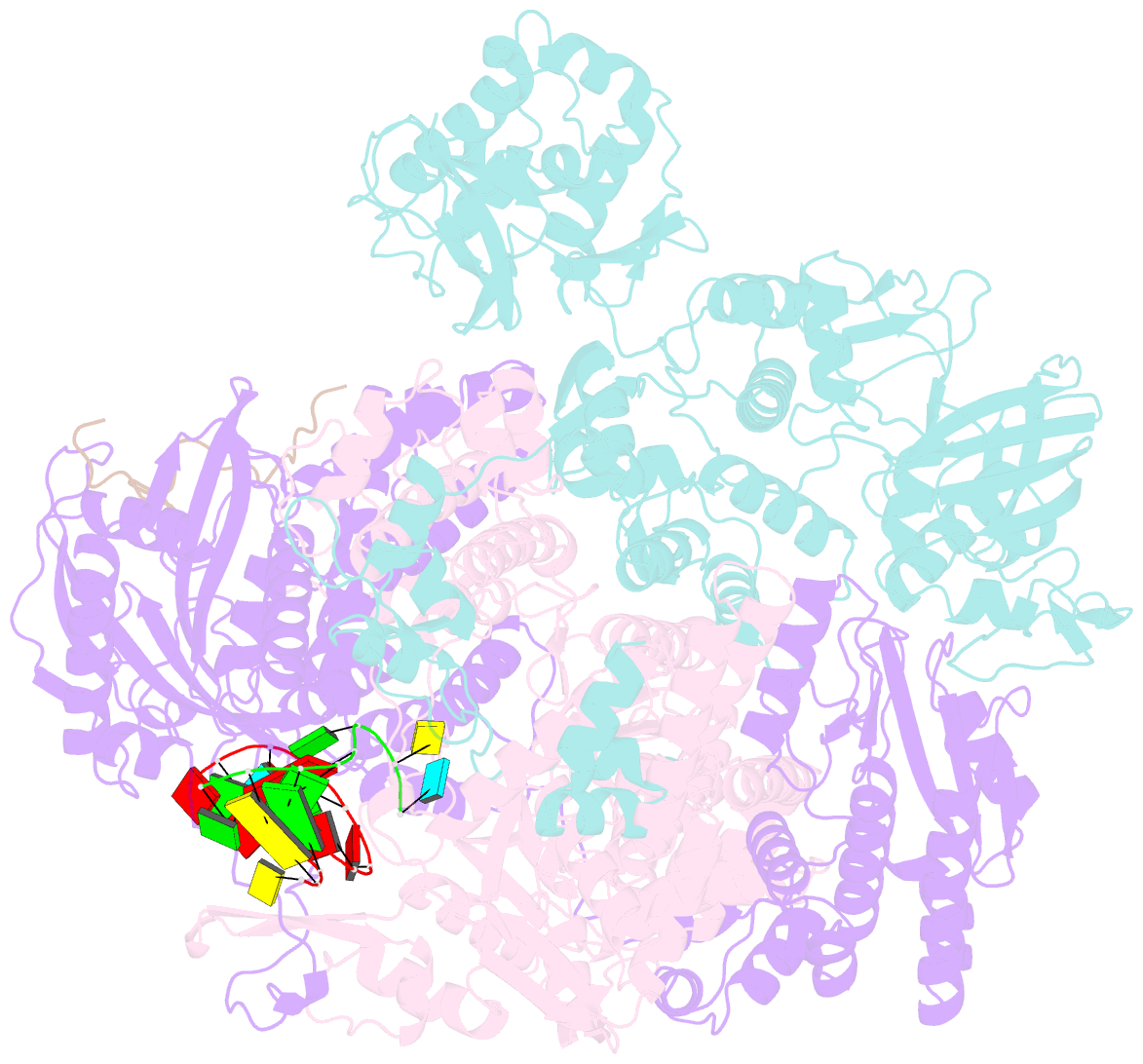
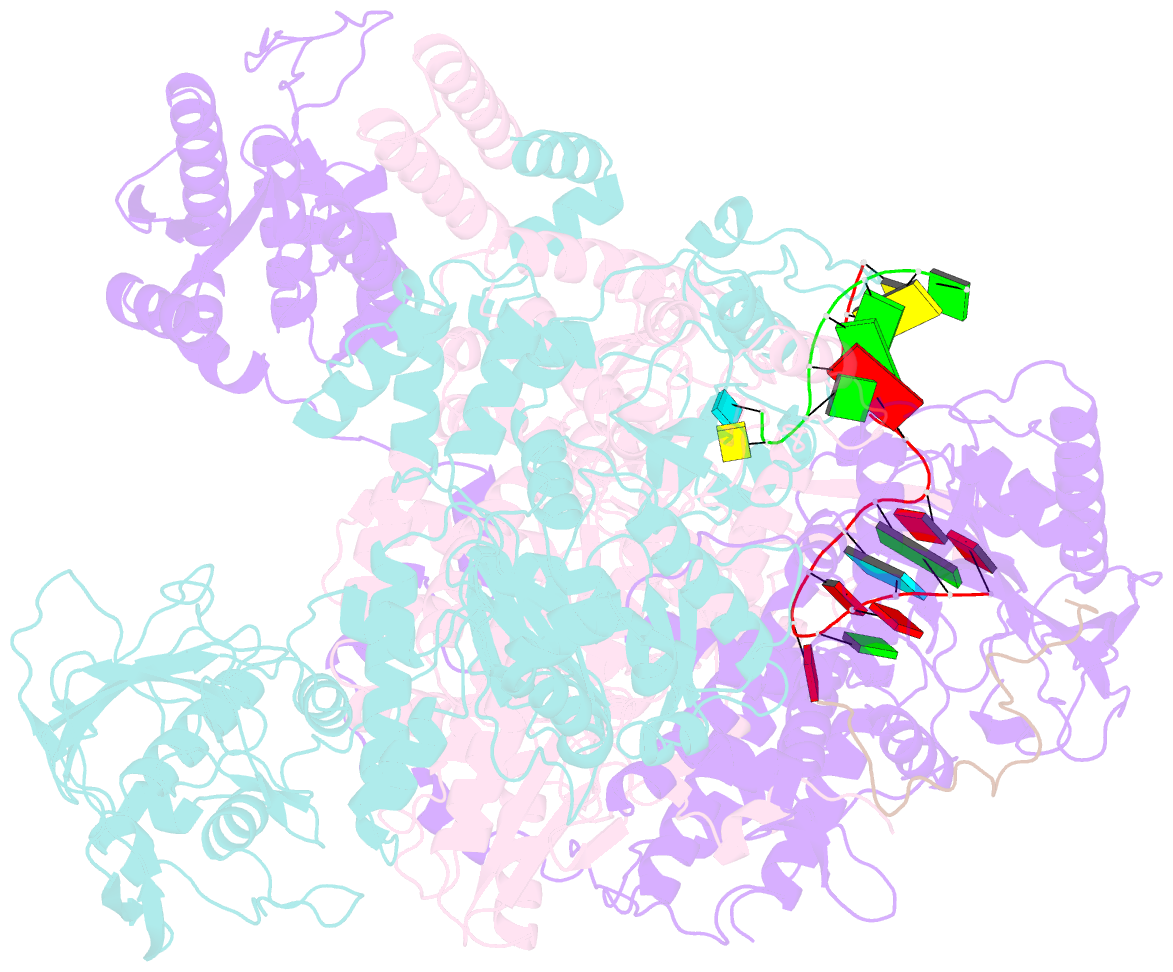
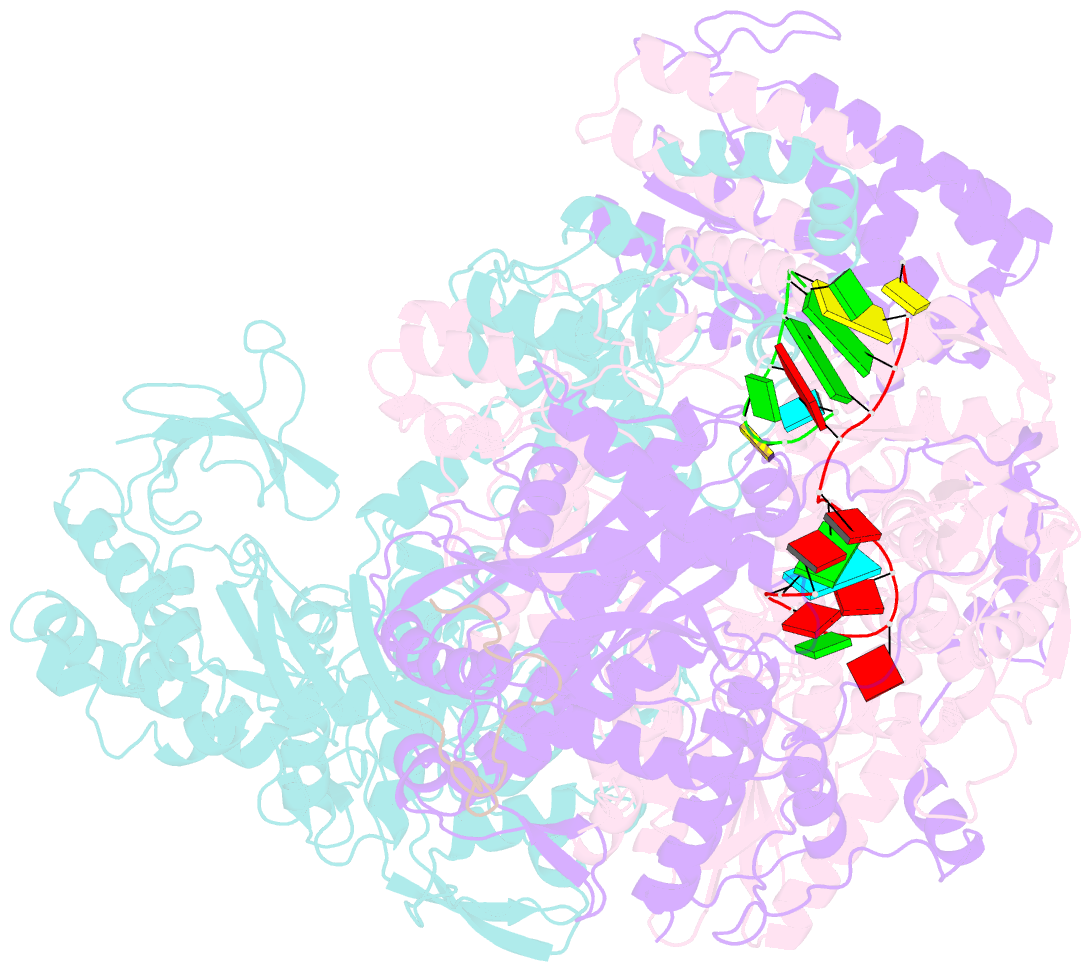
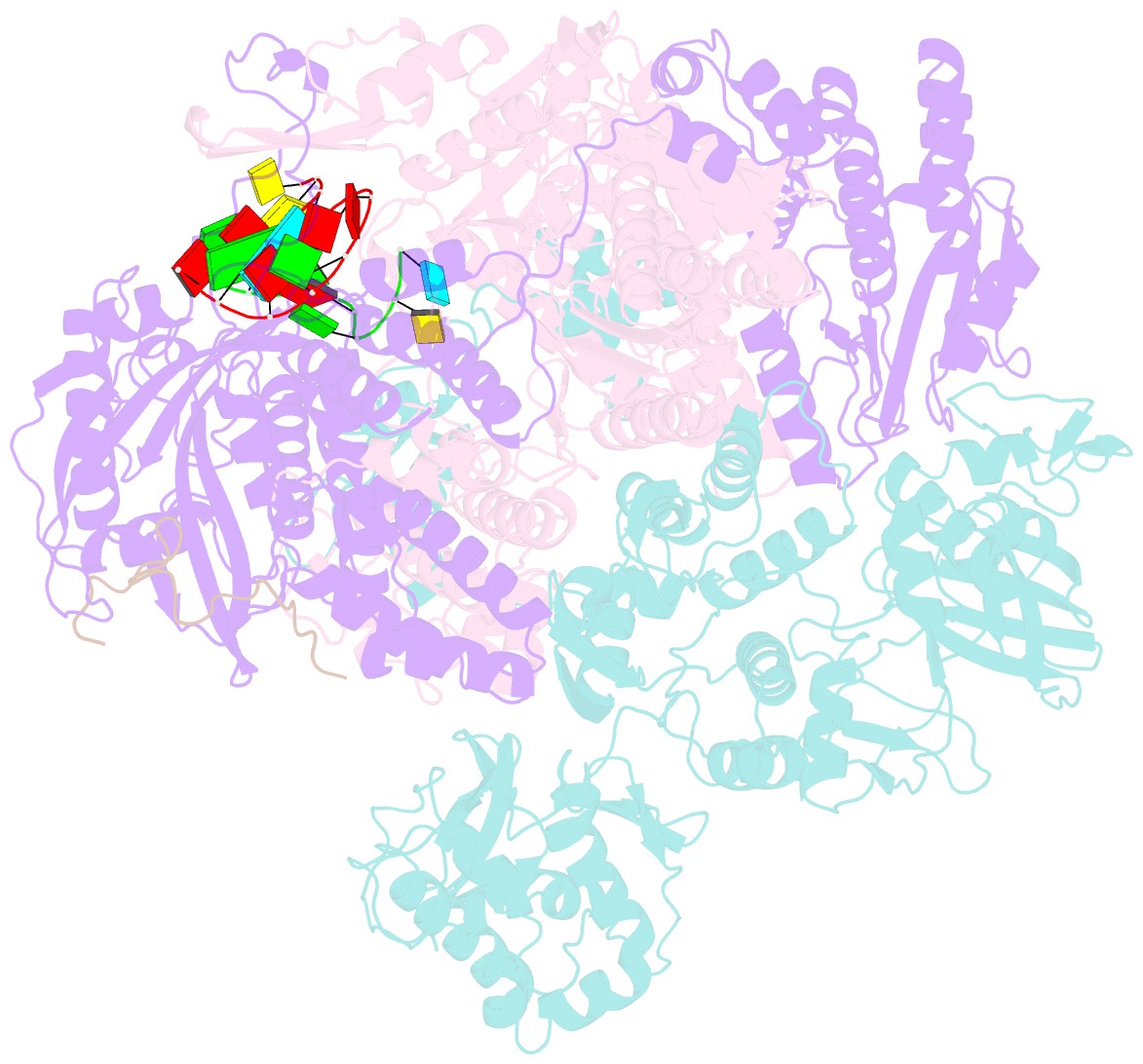
Summary information and primary citation
- PDB-id
- 8r65; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein
- Method
- cryo-EM (4.23 Å)
- Summary
- 1918 h1n1 viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated polii peptide with ordered pb2 c-terminal domains
- Reference
- Keown J, Baazaoui A, Sebesta M, Stefl R, Carrique L, Fodor E, Grimes JM (2024): "Structural and functional characterization of the interaction between the influenza A virus RNA polymerase and the CTD of host RNA polymerase II." J.Virol., 98, e0013824. doi: 10.1128/jvi.00138-24.
- Abstract
- Influenza A viruses, causing seasonal epidemics and occasional pandemics, rely on interactions with host proteins for their RNA genome transcription and replication. The viral RNA polymerase utilizes host RNA polymerase II (Pol II) and interacts with the serine 5 phosphorylated (pS5) C-terminal domain (CTD) of Pol II to initiate transcription. Our study, using single-particle electron cryomicroscopy (cryo-EM), reveals the structure of the 1918 pandemic influenza A virus polymerase bound to a synthetic pS5 CTD peptide composed of four heptad repeats mimicking the 52 heptad repeat mammalian Pol II CTD. The structure shows that the CTD peptide binds at the C-terminal domain of the PA viral polymerase subunit (PA-C) and reveals a previously unobserved position of the 627 domain of the PB2 subunit near the CTD. We identify crucial residues of the CTD peptide that mediate interactions with positively charged cavities on PA-C, explaining the preference of the viral polymerase for pS5 CTD. Functional analysis of mutants targeting the CTD-binding site within PA-C reveals reduced transcriptional function or defects in replication, highlighting the multifunctional role of PA-C in viral RNA synthesis. Our study provides insights into the structural and functional aspects of the influenza virus polymerase-host Pol II interaction and identifies a target for antiviral development.IMPORTANCEUnderstanding the intricate interactions between influenza A viruses and host proteins is crucial for developing targeted antiviral strategies. This study employs advanced imaging techniques to uncover the structural nuances of the 1918 pandemic influenza A virus polymerase bound to a specific host protein, shedding light on the vital process of viral RNA synthesis. The study identifies key amino acid residues in the influenza polymerase involved in binding host polymerase II (Pol II) and highlights their role in both viral transcription and genome replication. These findings not only deepen our understanding of the influenza virus life cycle but also pinpoint a potential target for antiviral development. By elucidating the structural and functional aspects of the influenza virus polymerase-host Pol II interaction, this research provides a foundation for designing interventions to disrupt viral replication and transcription, offering promising avenues for future antiviral therapies.