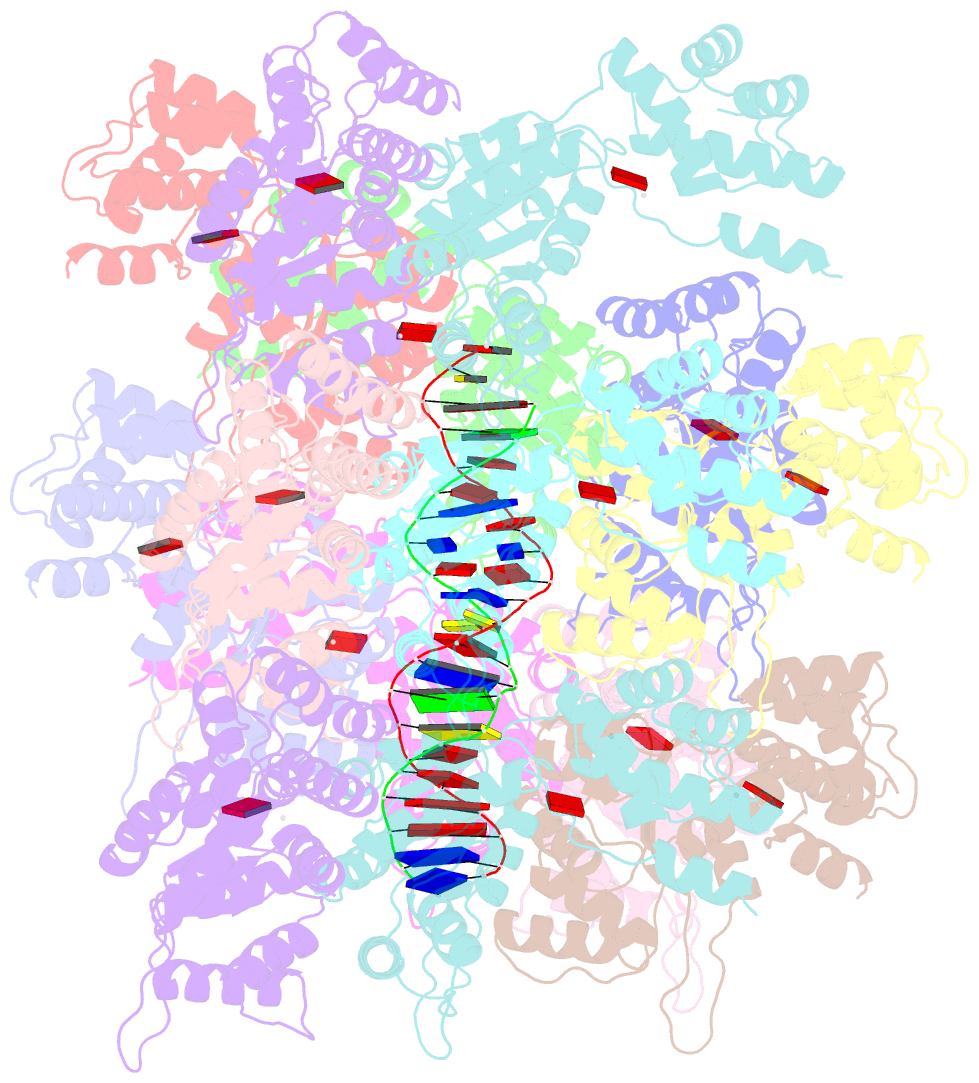
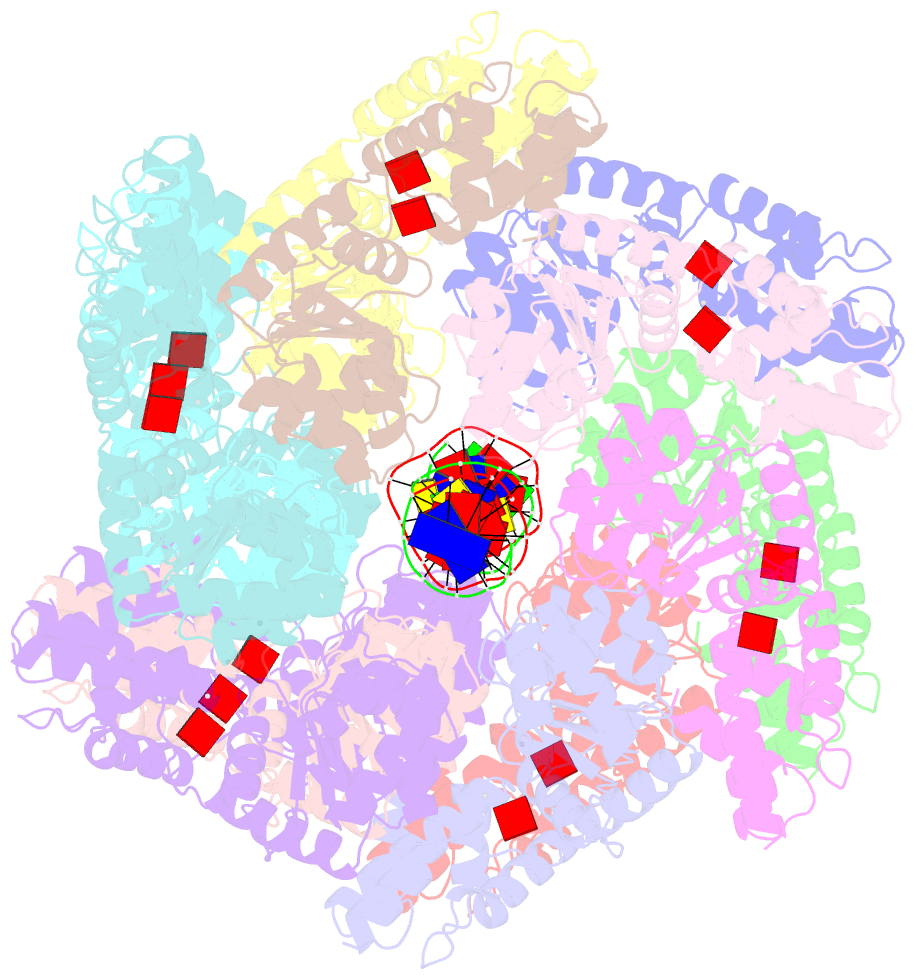
Summary information and primary citation
- PDB-id
- 8rku; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- cryo-EM (2.3 Å)
- Summary
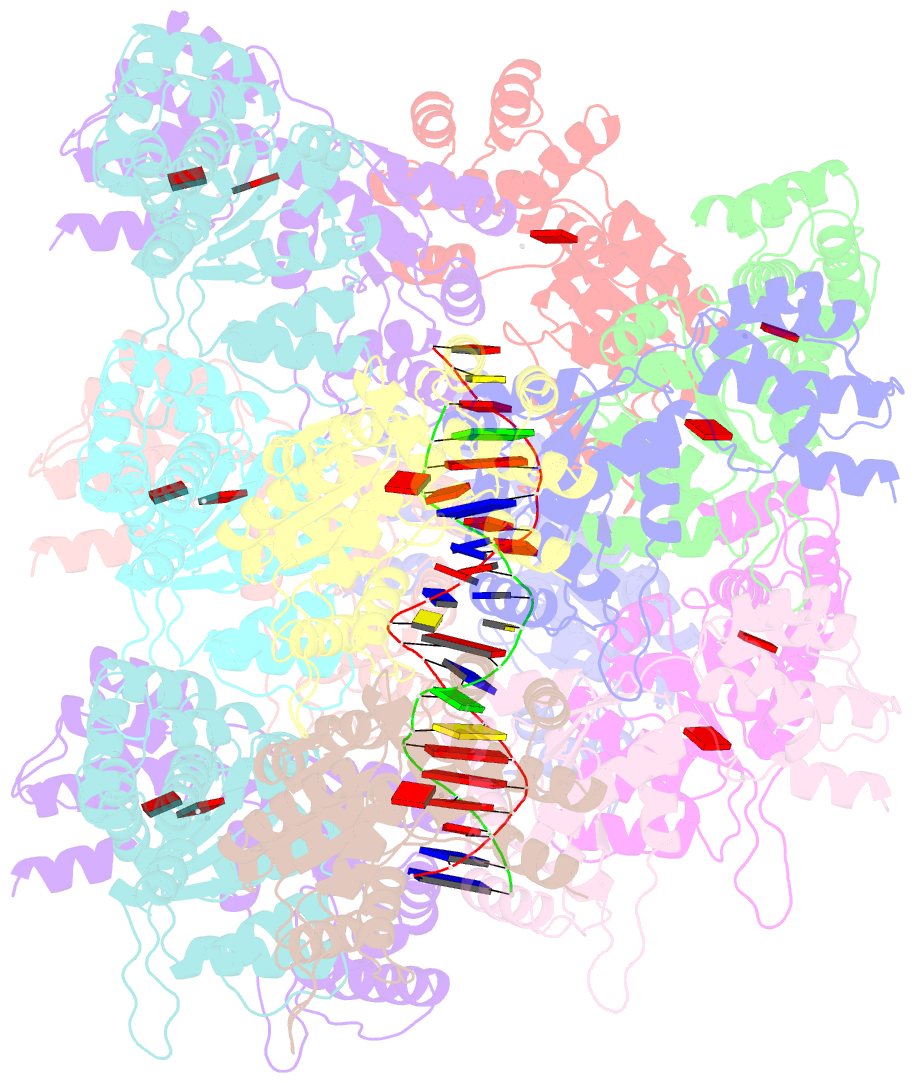
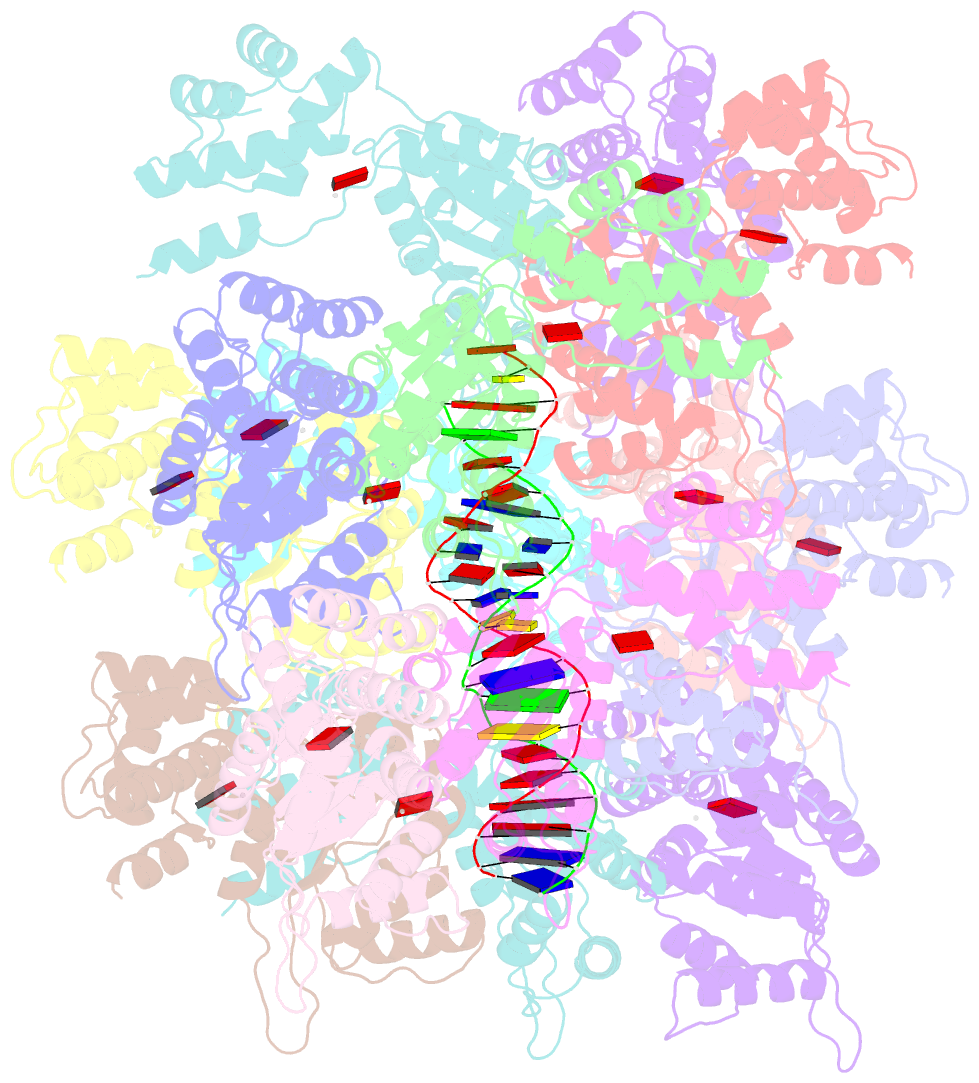
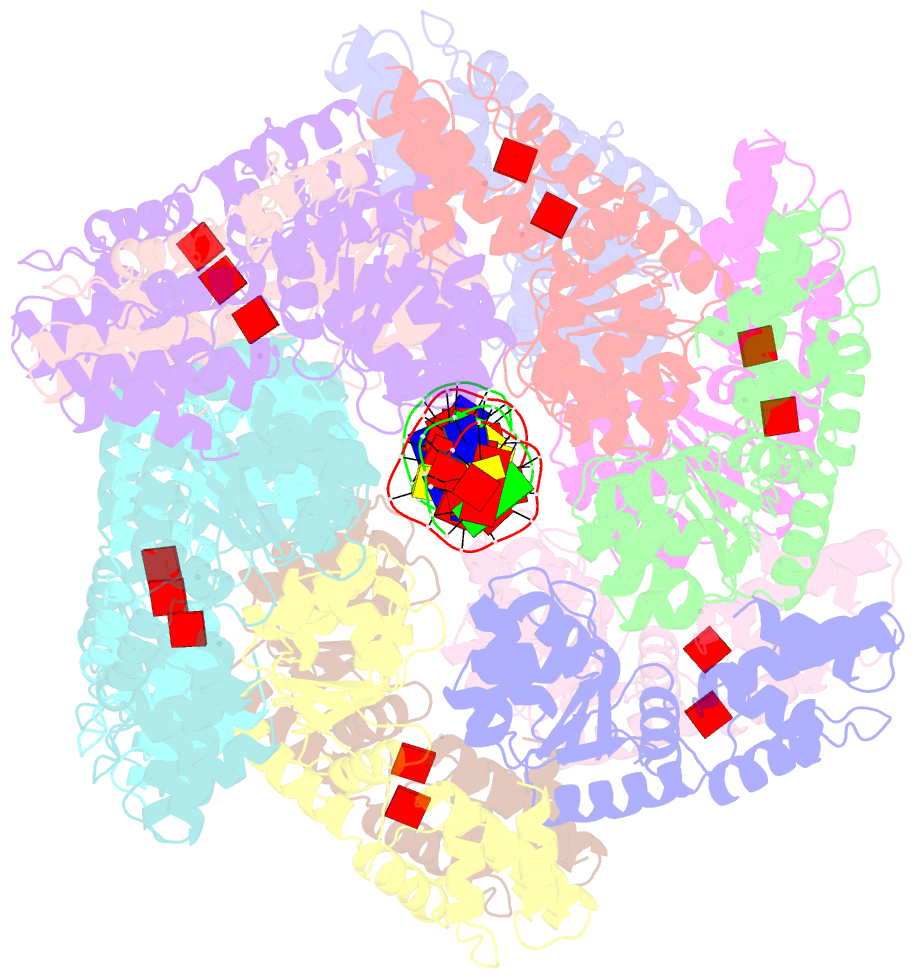
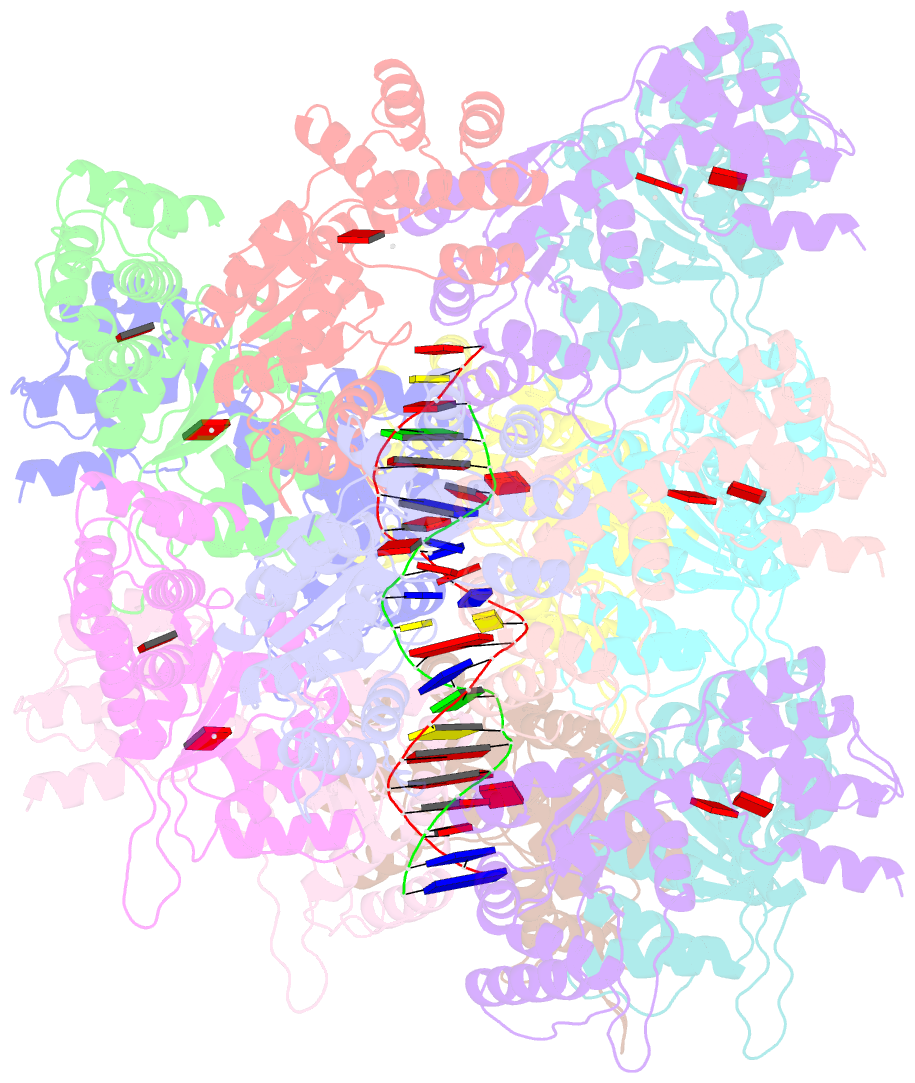
- Conformational landscape of the type v-k crispr-associated transposonintegration assembly cast v-k tnsc domain local-refinement map
- Reference
- Tenjo-Castano F, Sofos N, Stutzke LS, Temperini P, Fuglsang A, Pape T, Mesa P, Montoya G (2024): "Conformational landscape of the type V-K CRISPR-associated transposon integration assembly." Mol.Cell, 84, 2353. doi: 10.1016/j.molcel.2024.05.005.
- Abstract
- CRISPR-associated transposons (CASTs) are mobile genetic elements that co-opt CRISPR-Cas systems for RNA-guided DNA transposition. CASTs integrate large DNA cargos into the attachment (att) site independently of homology-directed repair and thus hold promise for eukaryotic genome engineering. However, the functional diversity and complexity of CASTs hinder an understanding of their mechanisms. Here, we present the high-resolution cryoelectron microscopy (cryo-EM) structure of the reconstituted ∼1 MDa post-transposition complex of the type V-K CAST, together with different assembly intermediates and diverse TnsC filament lengths, thus enabling the recapitulation of the integration complex formation. The results of mutagenesis experiments probing the roles of specific residues and TnsB-binding sites show that transposition activity can be enhanced and suggest that the distance between the PAM and att sites is determined by the lengths of the TnsB C terminus and the TnsC filament. This singular model of RNA-guided transposition provides a foundation for repurposing the system for genome-editing applications.