Summary information and primary citation
- PDB-id
- 8sny; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- cryo-EM (3.41 Å)
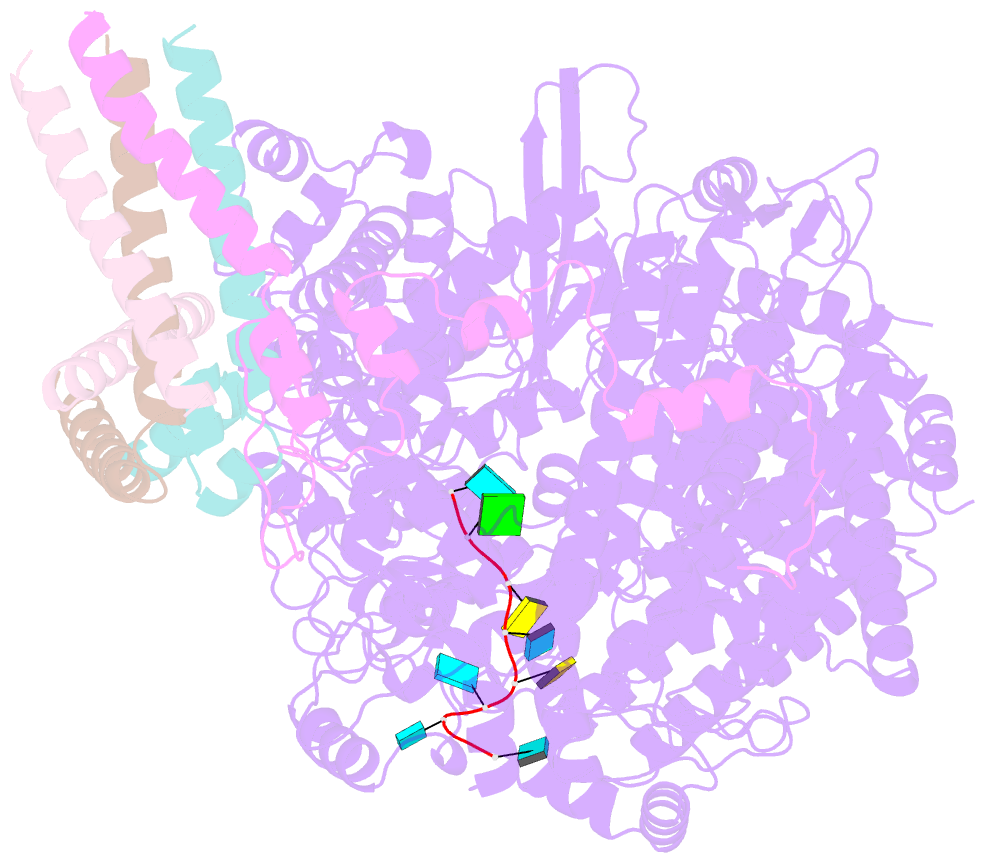
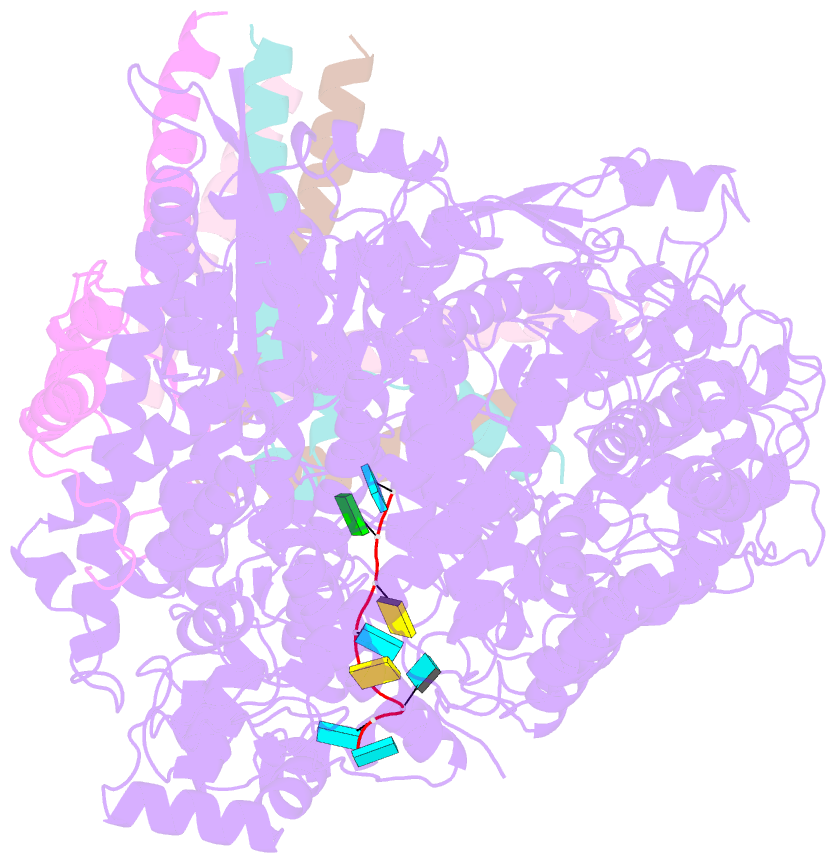
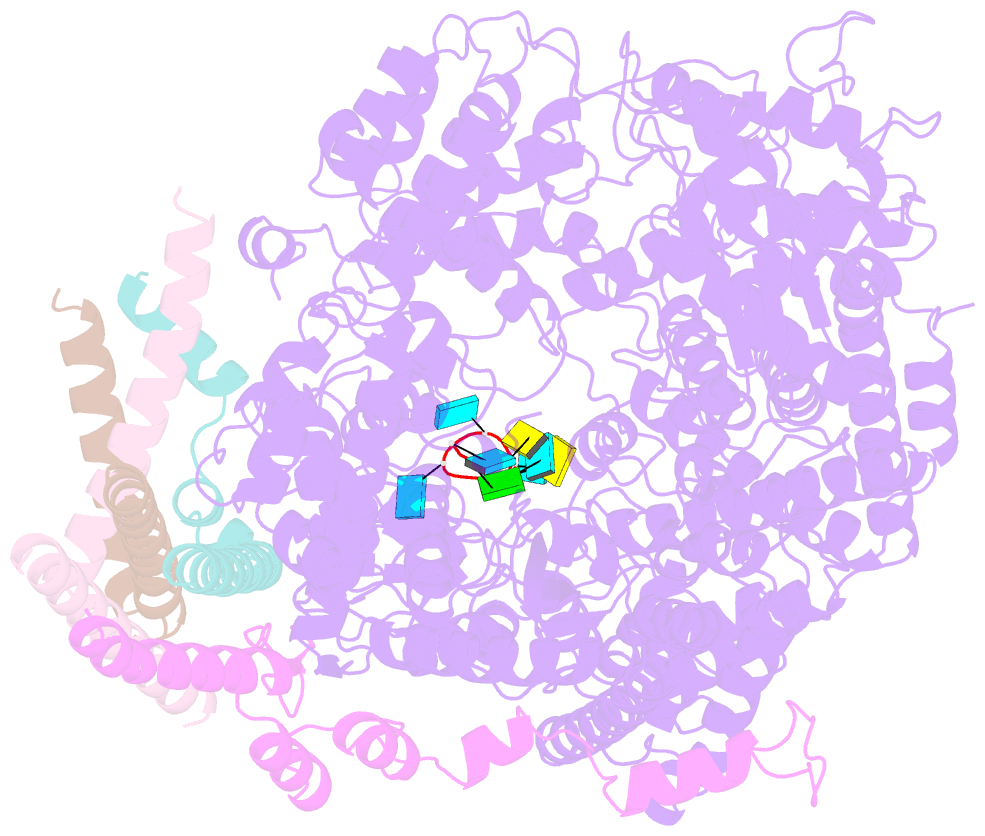
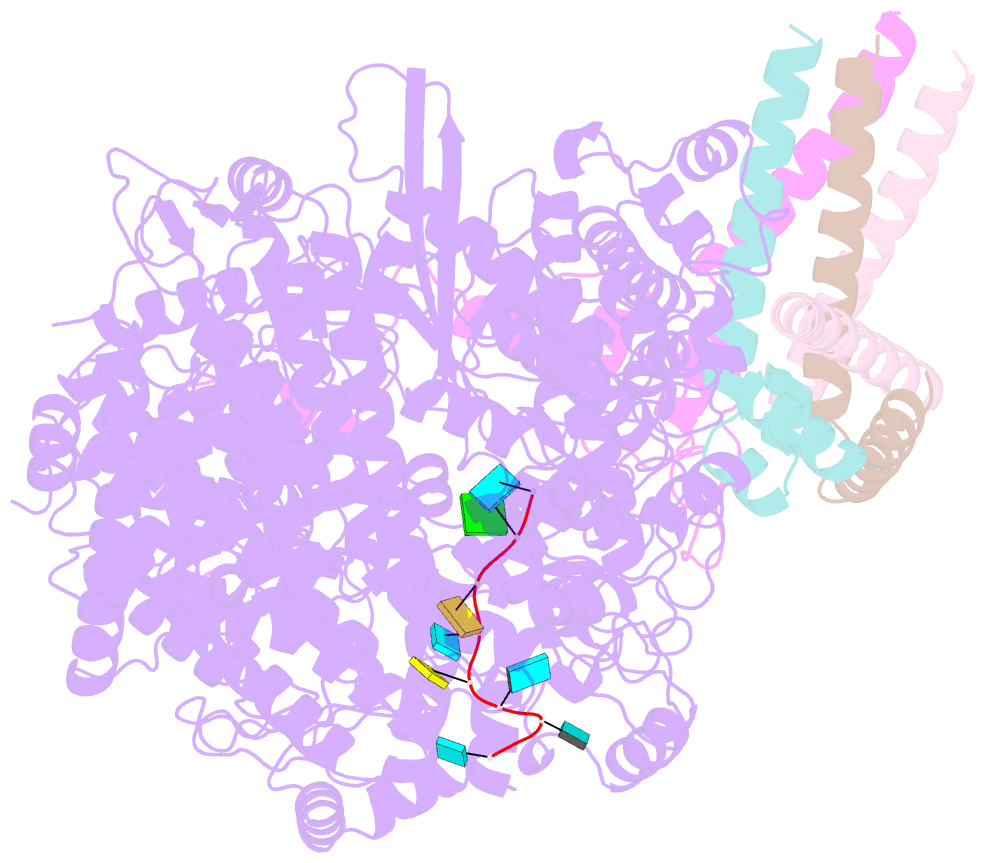
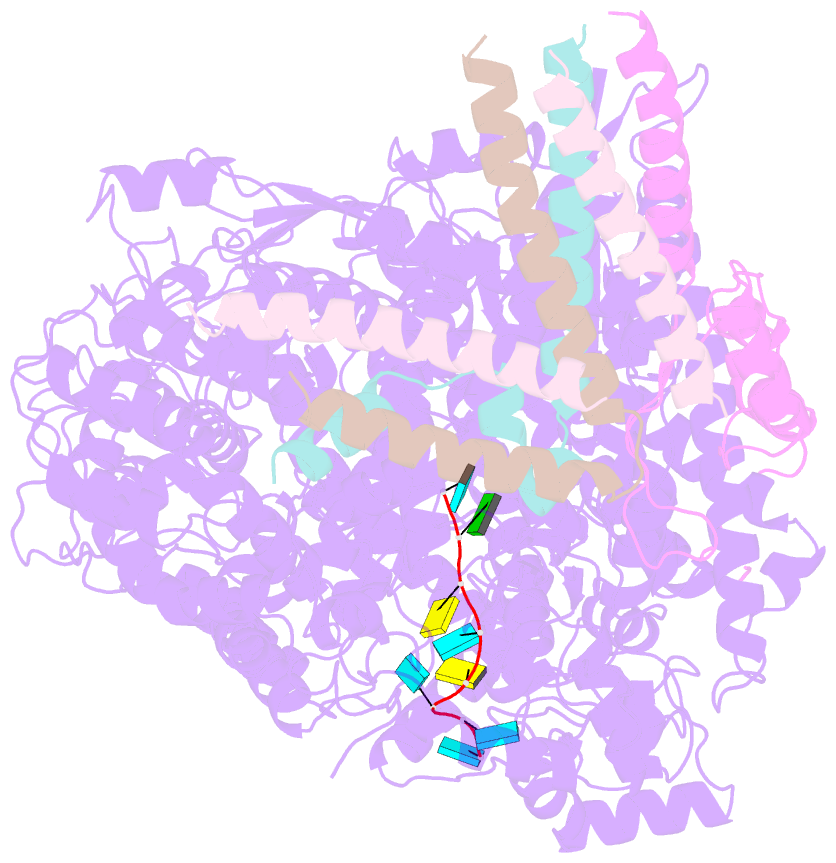
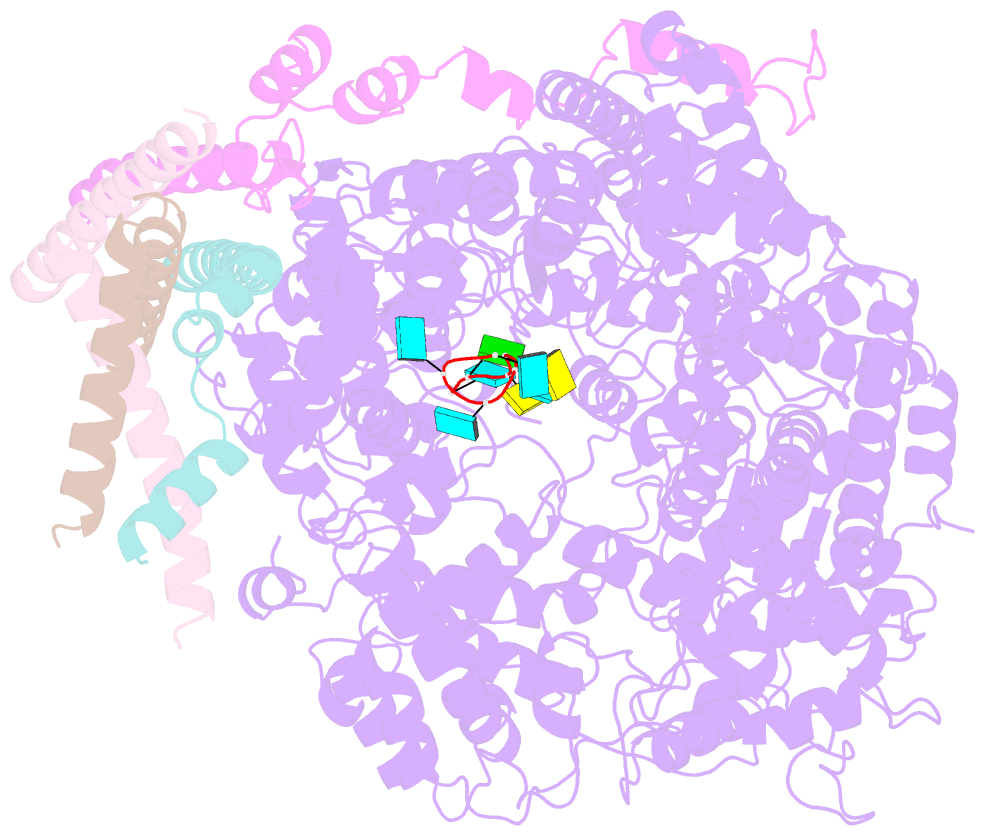
- Summary
- cryo-EM structure of the respiratory syncytial virus polymerase (l:p) bound to the trailer complementary promoter
- Reference
- Cao D, Gao Y, Chen Z, Gooneratne I, Roesler C, Mera C, D'Cunha P, Antonova A, Katta D, Romanelli S, Wang Q, Rice S, Lemons W, Ramanathan A, Liang B (2024): "Structures of the promoter-bound respiratory syncytial virus polymerase." Nature, 625, 611-617. doi: 10.1038/s41586-023-06867-y.
- Abstract
- The respiratory syncytial virus (RSV) polymerase is a multifunctional RNA-dependent RNA polymerase composed of the large (L) protein and the phosphoprotein (P). It transcribes the RNA genome into ten viral mRNAs and replicates full-length viral genomic and antigenomic RNAs1. The RSV polymerase initiates RNA synthesis by binding to the conserved 3'-terminal RNA promoters of the genome or antigenome2. However, the lack of a structure of the RSV polymerase bound to the RNA promoter has impeded the mechanistic understanding of RSV RNA synthesis. Here we report cryogenic electron microscopy structures of the RSV polymerase bound to its genomic and antigenomic viral RNA promoters, representing two of the first structures of an RNA-dependent RNA polymerase in complex with its RNA promoters in non-segmented negative-sense RNA viruses. The overall structures of the promoter-bound RSV polymerases are similar to that of the unbound (apo) polymerase. Our structures illustrate the interactions between the RSV polymerase and the RNA promoters and provide the structural basis for the initiation of RNA synthesis at positions 1 and 3 of the RSV promoters. These structures offer a deeper understanding of the pre-initiation state of the RSV polymerase and could aid in antiviral research against RSV.