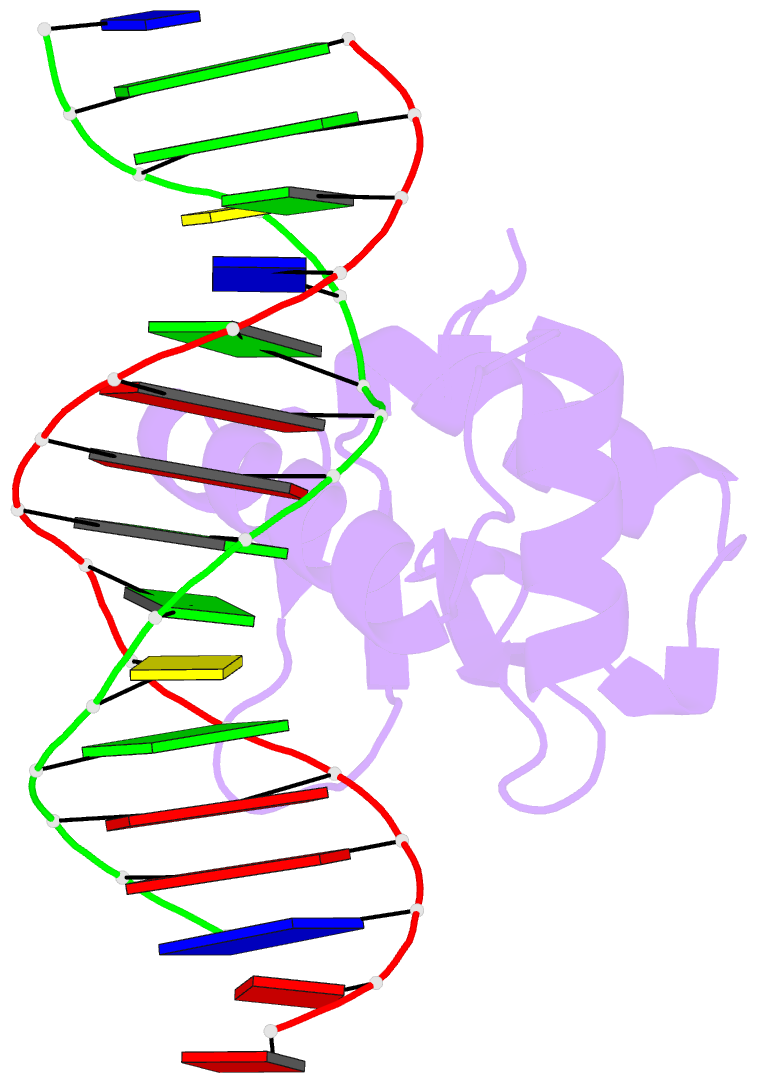
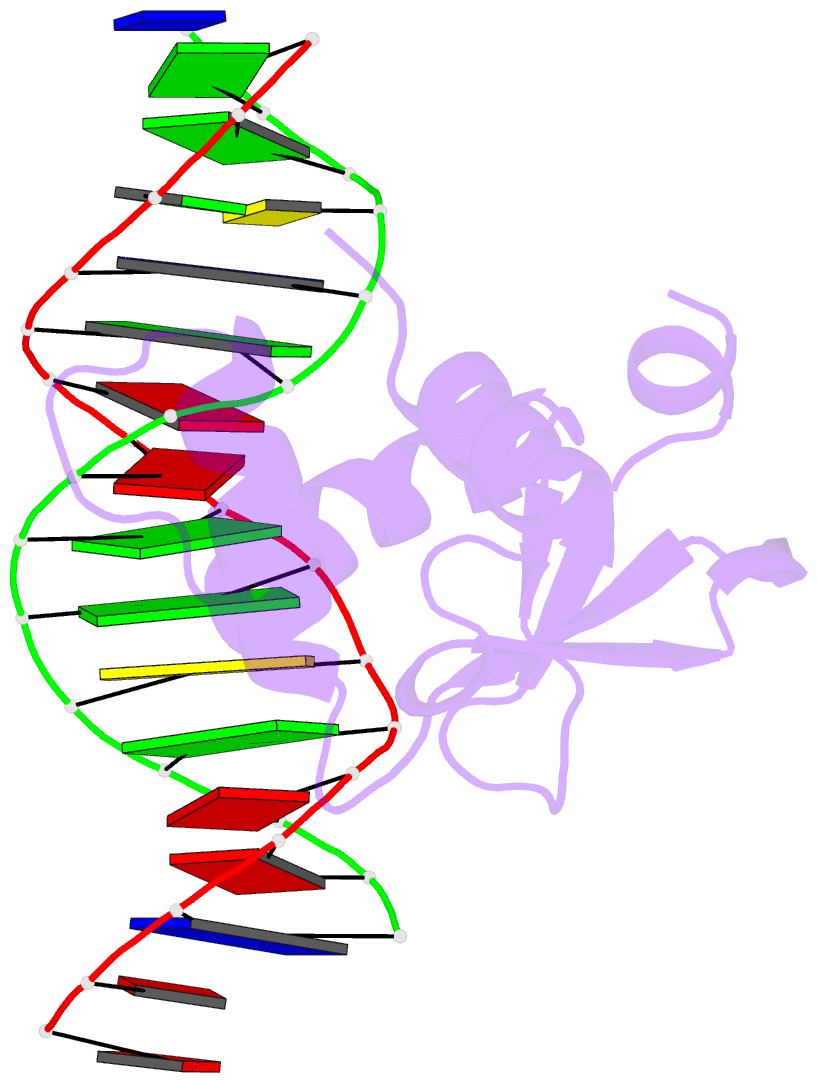
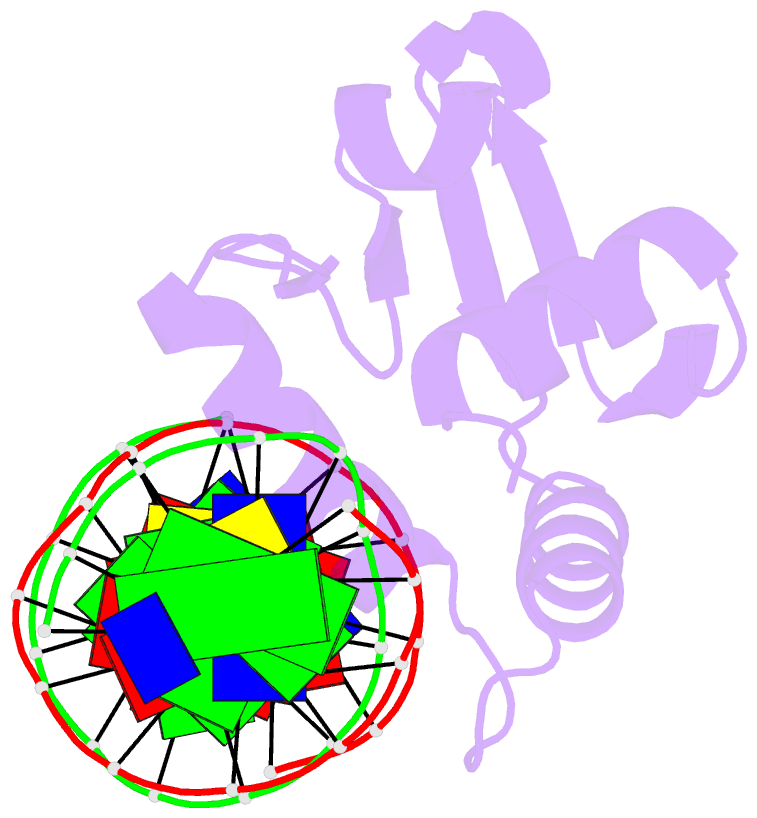
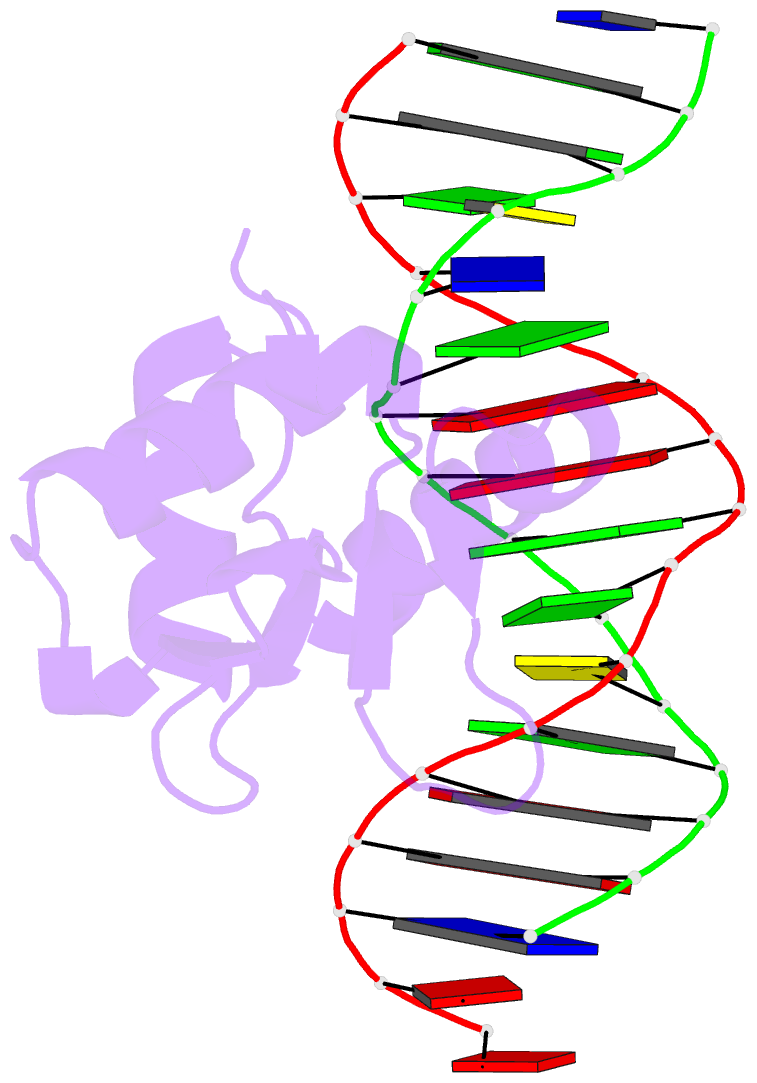
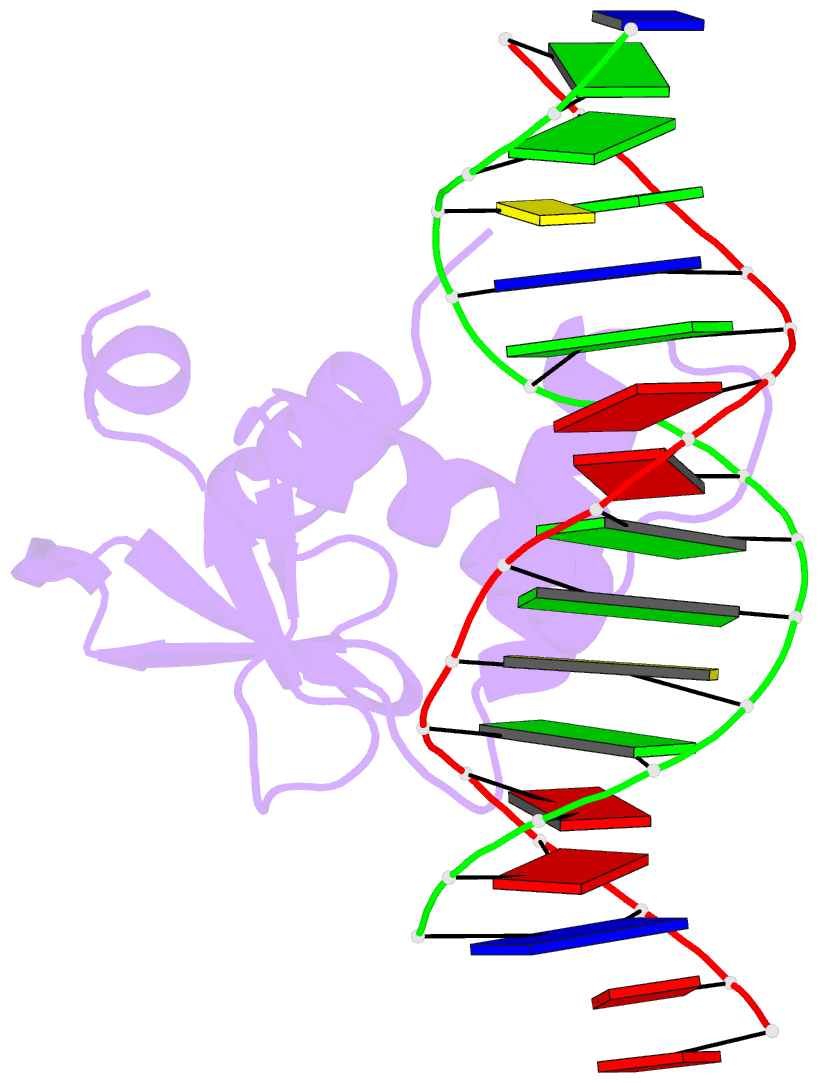
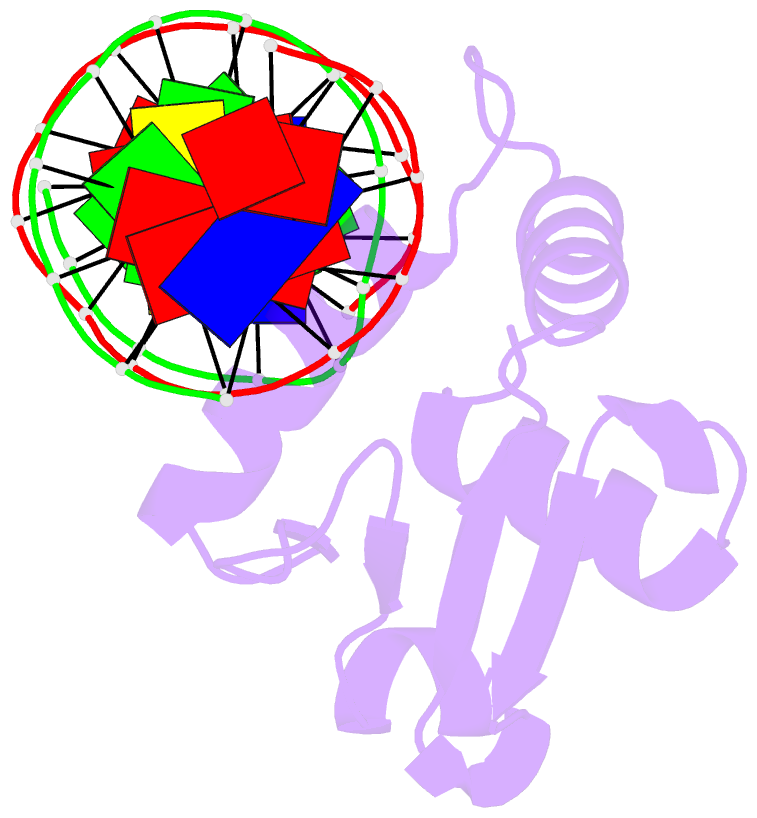
Summary information and primary citation
- PDB-id
- 8sp1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.62 Å)
- Summary
- Chimeric ets-domain of murine pu.1 harboring the corresponding beta-strand 3 (s3) residues from murine ets-1 in complex with d(aataagcgiaagtggg)
- Reference
- Vernon TN, Terrell JR, Albrecht AV, Germann MW, Wilson WD, Poon GMK (2024): "Dissection of integrated readout reveals the structural thermodynamics of DNA selection by transcription factors." Structure, 32, 83-96.e4. doi: 10.1016/j.str.2023.11.003.
- Abstract
- Nucleobases such as inosine have been extensively utilized to map direct contacts by proteins in the DNA groove. Their deployment as targeted probes of dynamics and hydration, which are dominant thermodynamic drivers of affinity and specificity, has been limited by a paucity of suitable experimental models. We report a joint crystallographic, thermodynamic, and computational study of the bidentate complex of the arginine side chain with a Watson-Crick guanine (Arg×GC), a highly specific configuration adopted by major transcription factors throughout the eukaryotic branches in the Tree of Life. Using the ETS-family factor PU.1 as a high-resolution structural framework, inosine substitution for guanine resulted in a sharp dissection of conformational dynamics and hydration and elucidated their role in the DNA specificity of PU.1. Our work suggests an under-exploited utility of modified nucleobases in untangling the structural thermodynamics of interactions, such as the Arg×GC motif, where direct and indirect readout are tightly integrated.