Summary information and primary citation
- PDB-id
- 8sss; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.3 Å)
- Summary
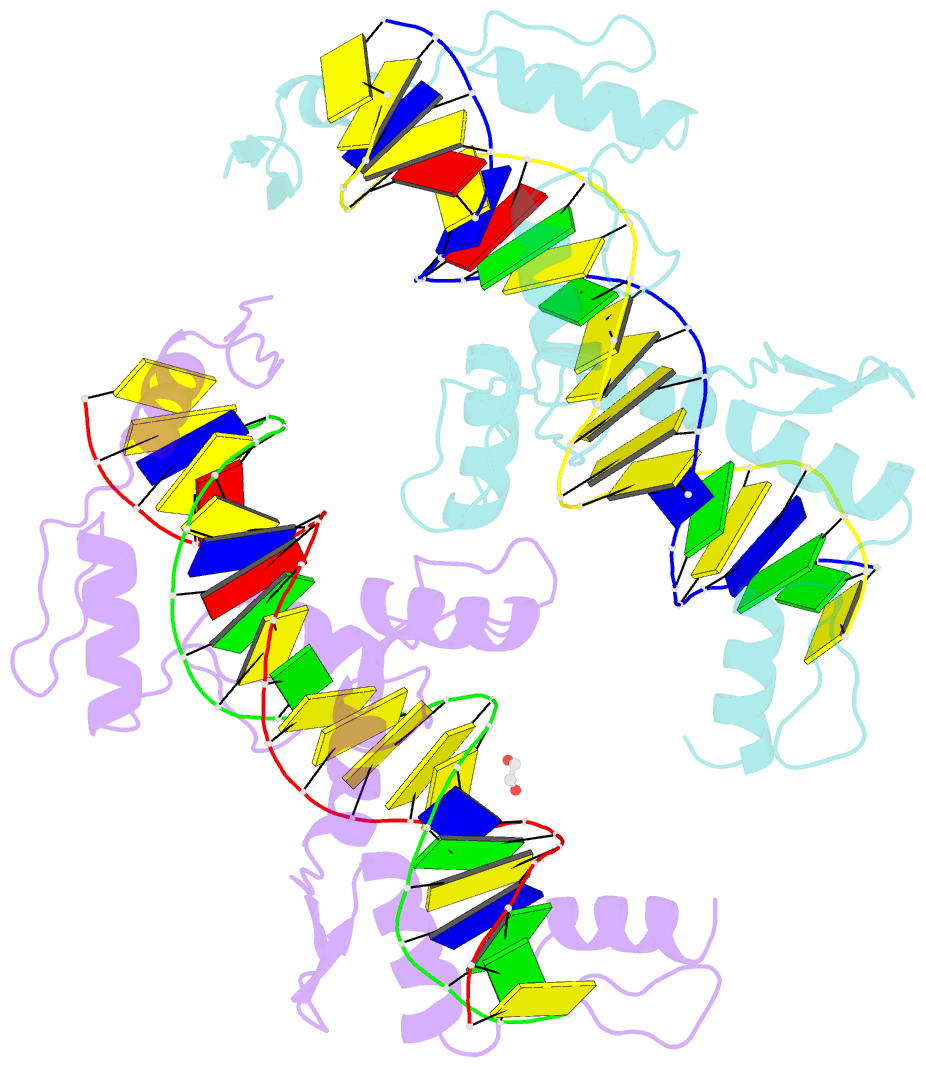
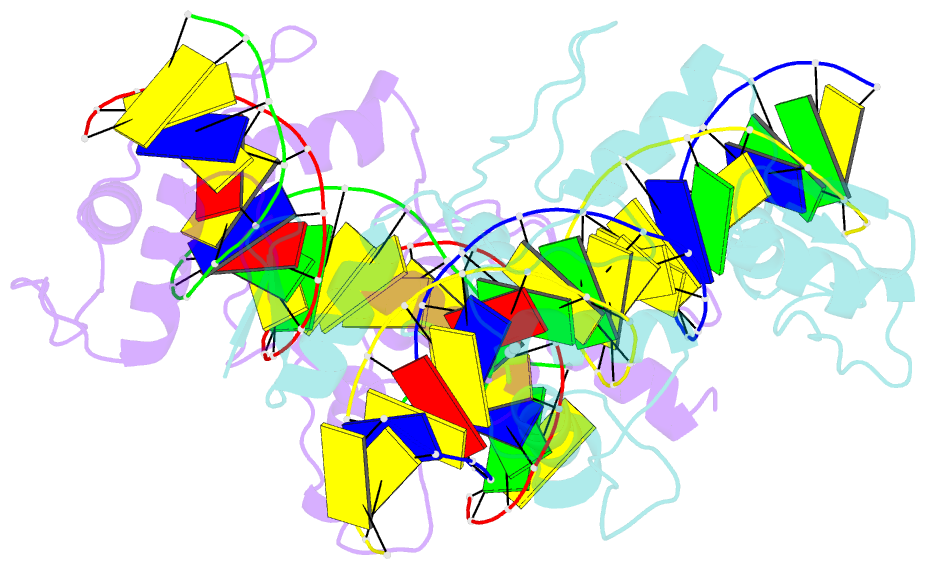
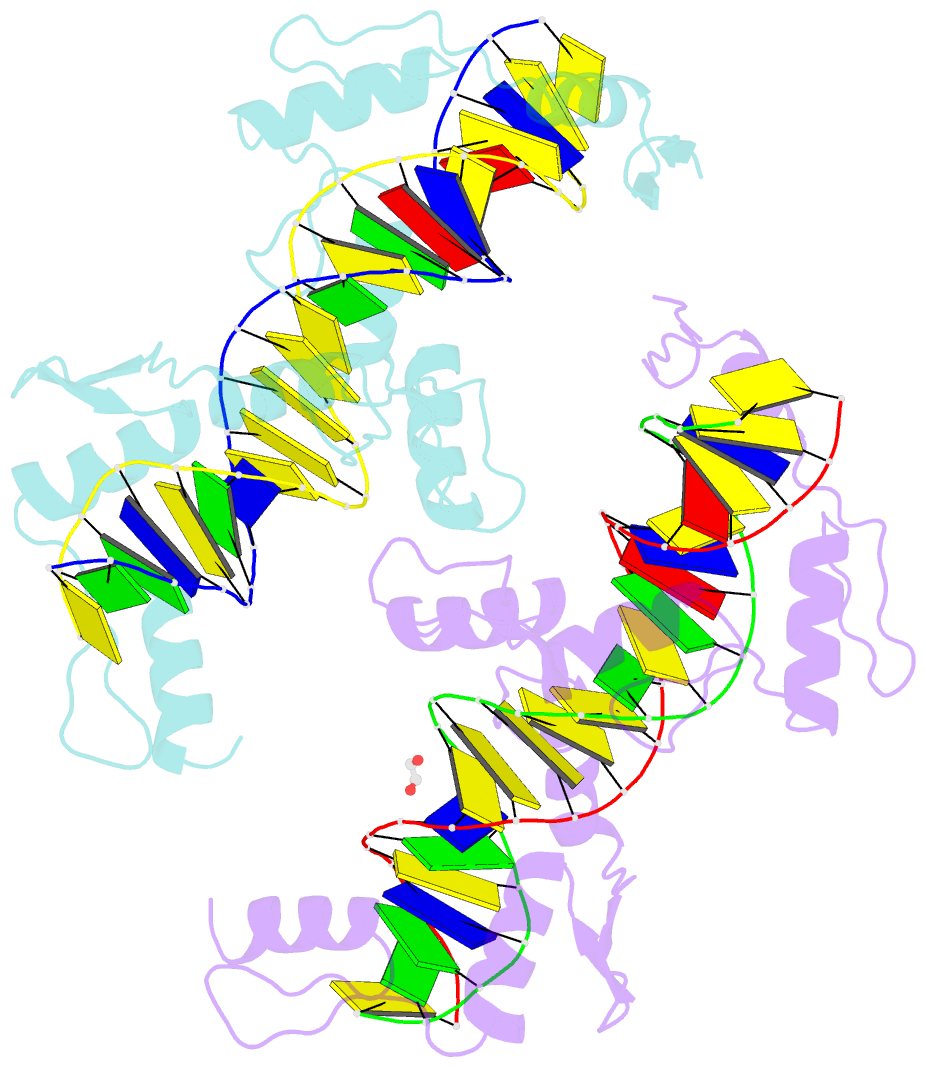
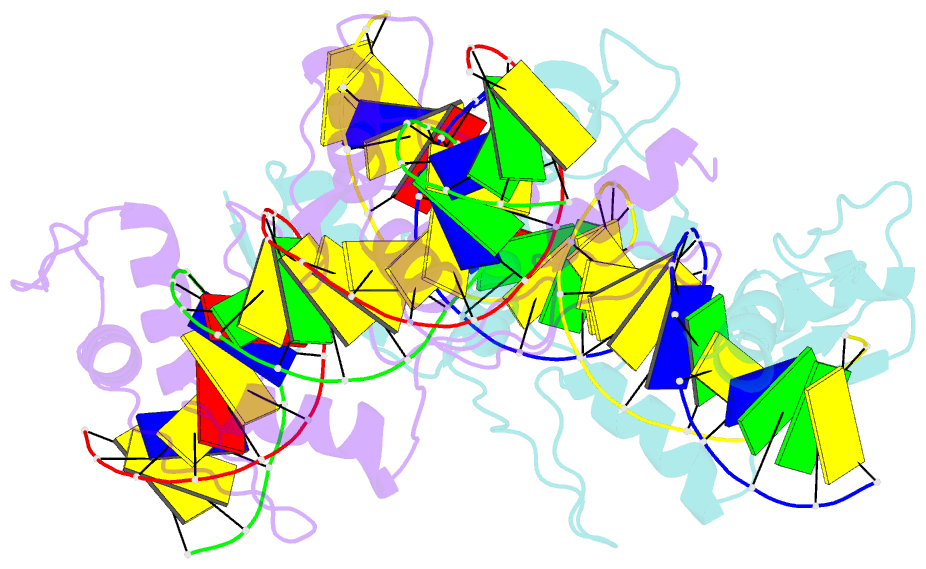
- Znfs 1-7 of ccctc-binding factor (ctcf) complexed with 23mer
- Reference
- Yang J, Horton JR, Liu B, Corces VG, Blumenthal RM, Zhang X, Cheng X (2023): "Structures of CTCF-DNA complexes including all 11 zinc fingers." Nucleic Acids Res., 51, 8447-8462. doi: 10.1093/nar/gkad594.
- Abstract
- The CCCTC-binding factor (CTCF) binds tens of thousands of enhancers and promoters on mammalian chromosomes by means of its 11 tandem zinc finger (ZF) DNA-binding domain. In addition to the 12-15-bp CORE sequence, some of the CTCF binding sites contain 5' upstream and/or 3' downstream motifs. Here, we describe two structures for overlapping portions of human CTCF, respectively, including ZF1-ZF7 and ZF3-ZF11 in complex with DNA that incorporates the CORE sequence together with either 3' downstream or 5' upstream motifs. Like conventional tandem ZF array proteins, ZF1-ZF7 follow the right-handed twist of the DNA, with each finger occupying and recognizing one triplet of three base pairs in the DNA major groove. ZF8 plays a unique role, acting as a spacer across the DNA minor groove and positioning ZF9-ZF11 to make cross-strand contacts with DNA. We ascribe the difference between the two subgroups of ZF1-ZF7 and ZF8-ZF11 to residues at the two positions -6 and -5 within each finger, with small residues for ZF1-ZF7 and bulkier and polar/charged residues for ZF8-ZF11. ZF8 is also uniquely rich in basic amino acids, which allows salt bridges to DNA phosphates in the minor groove. Highly specific arginine-guanine and glutamine-adenine interactions, used to recognize G:C or A:T base pairs at conventional base-interacting positions of ZFs, also apply to the cross-strand interactions adopted by ZF9-ZF11. The differences between ZF1-ZF7 and ZF8-ZF11 can be rationalized structurally and may contribute to recognition of high-affinity CTCF binding sites.