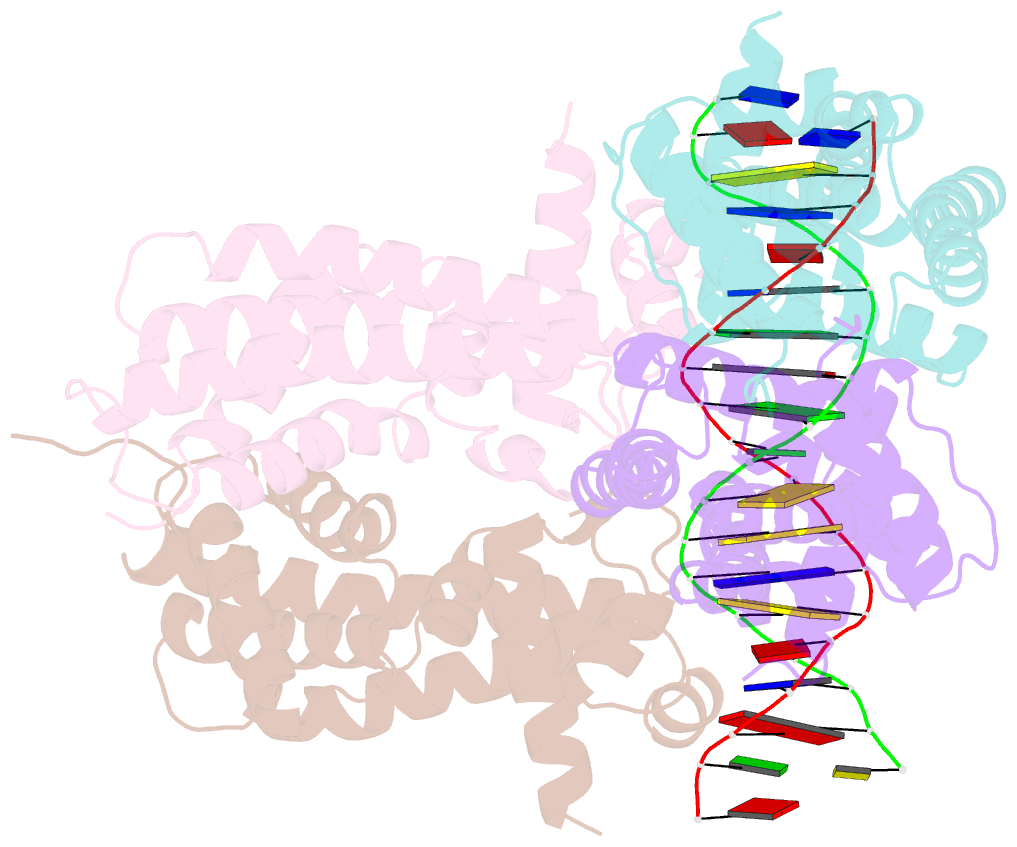
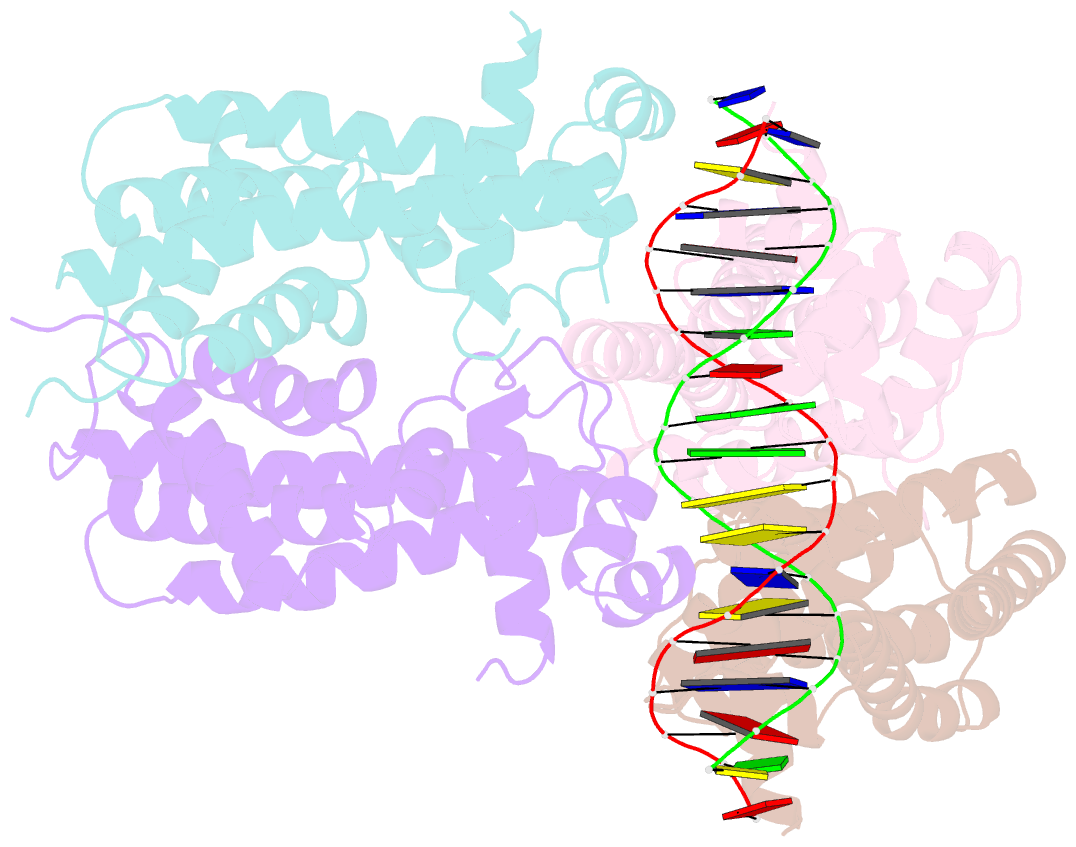
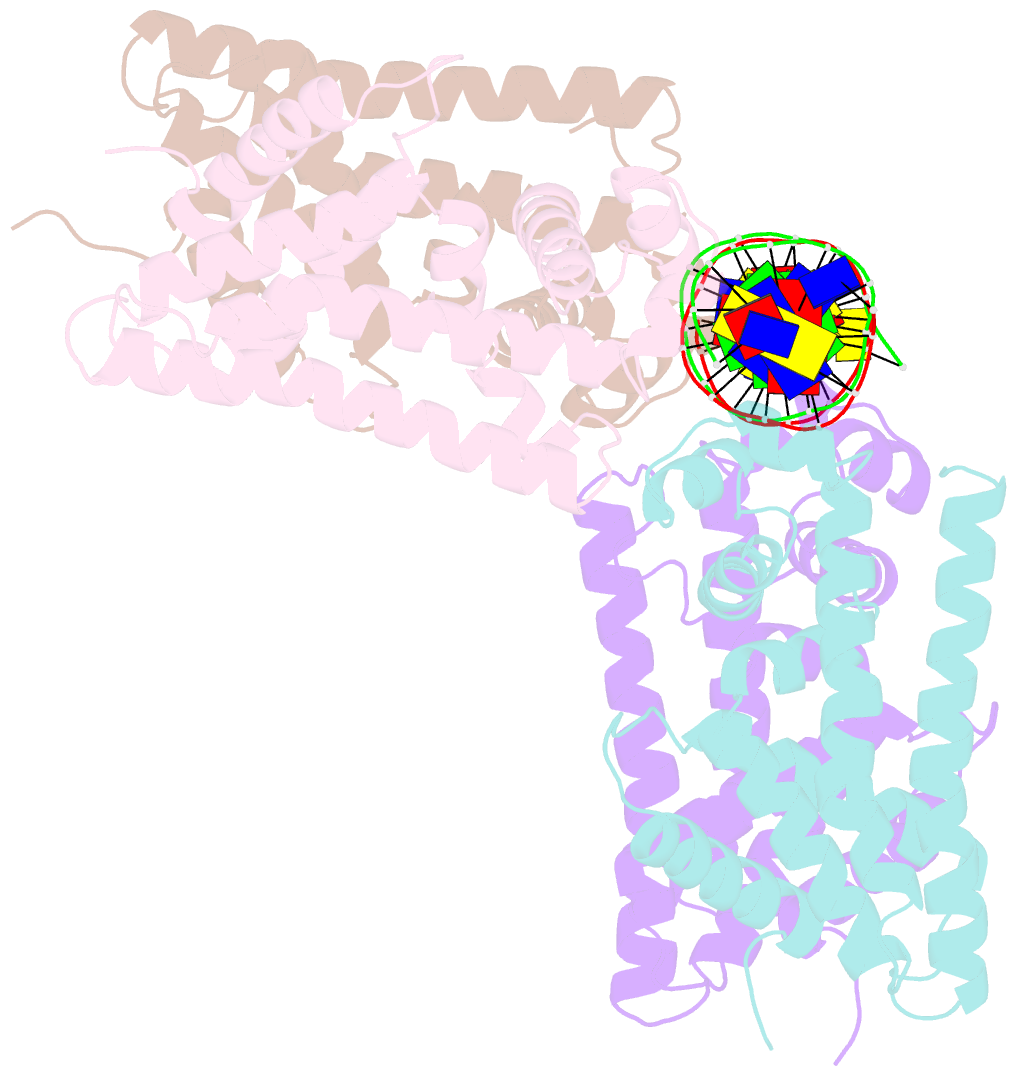
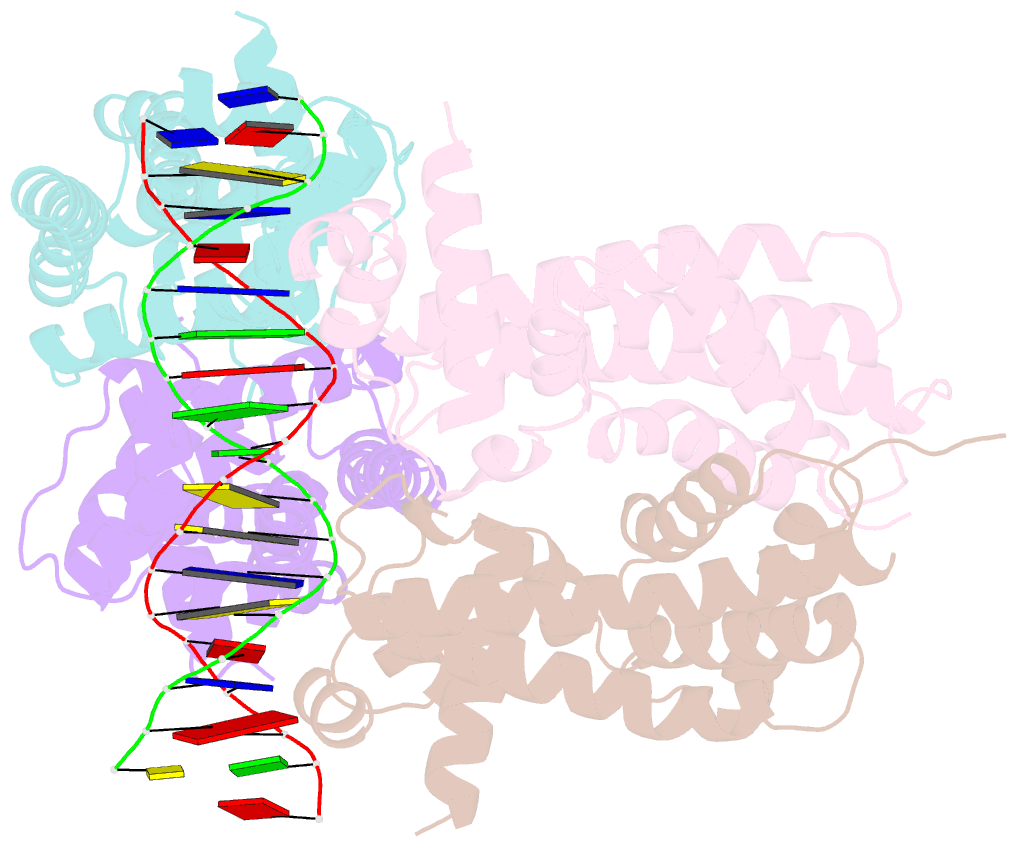
Summary information and primary citation
- PDB-id
- 8sva; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.96 Å)
- Summary
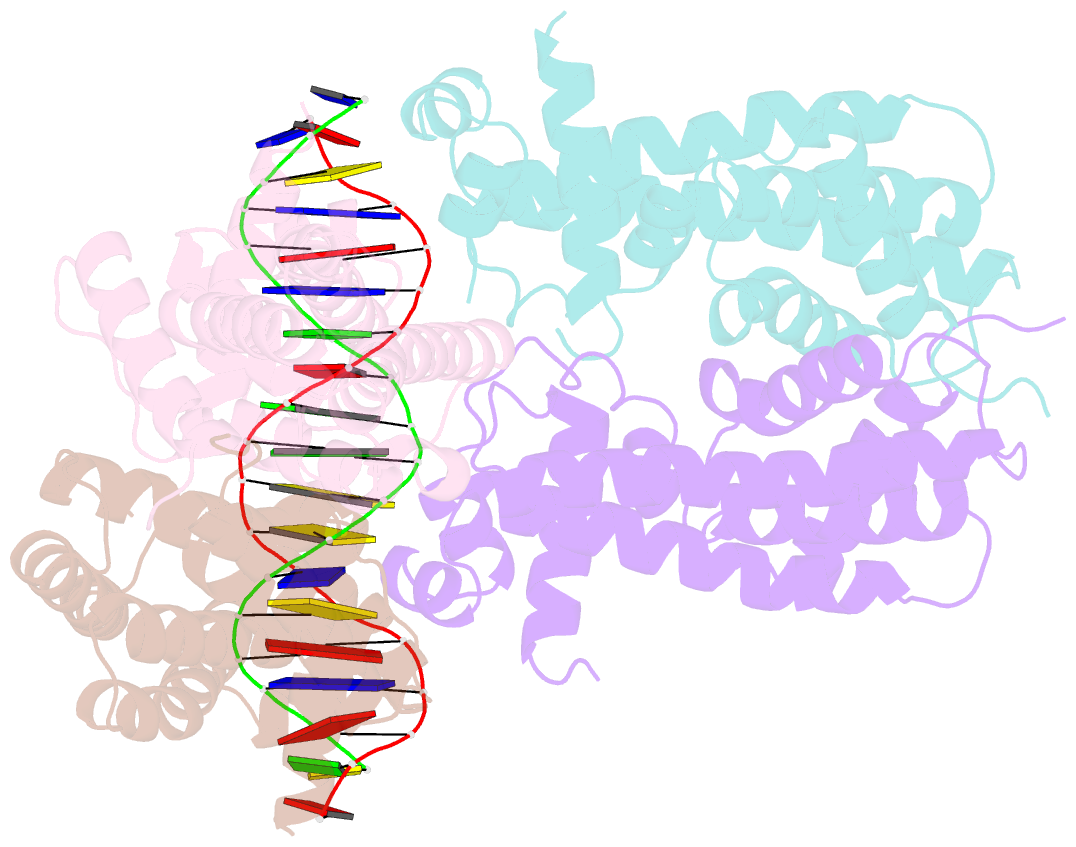
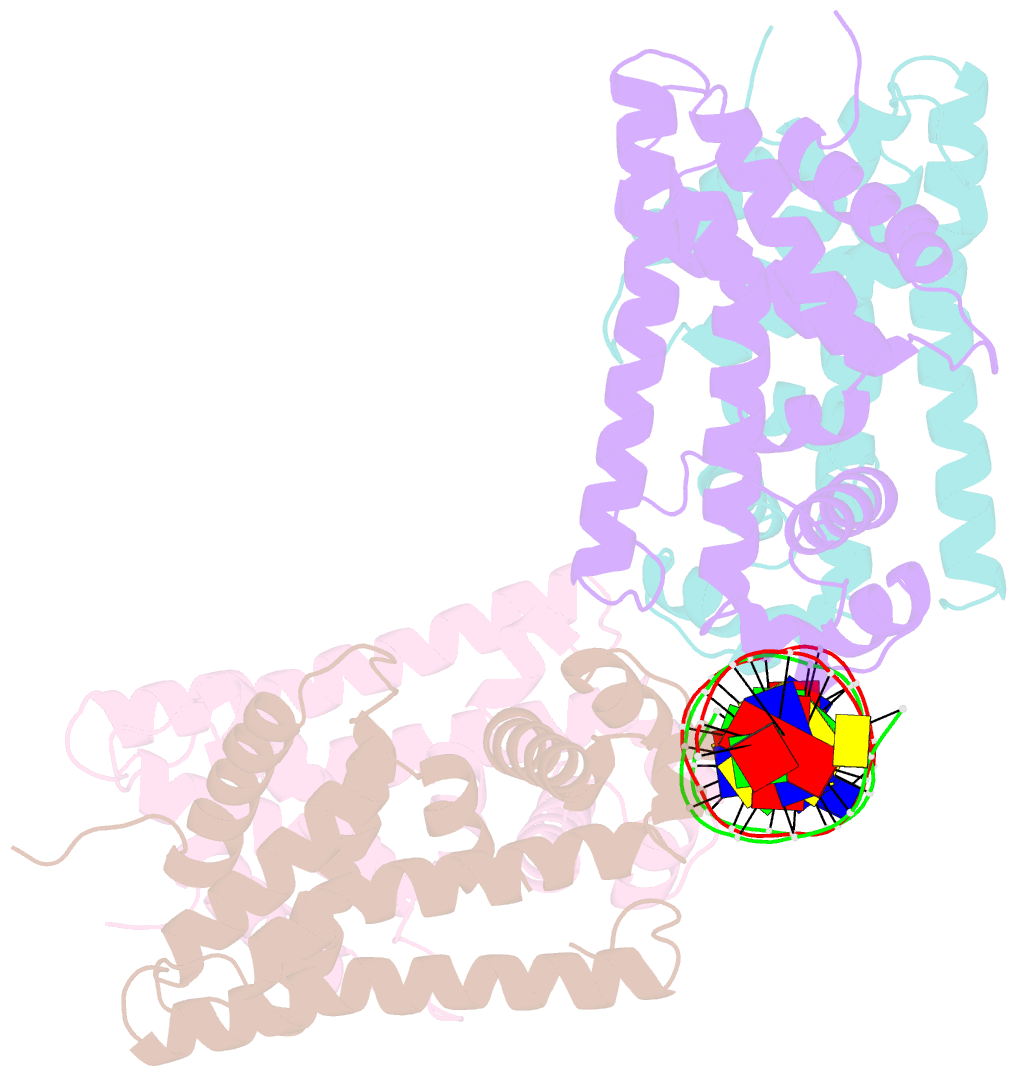
- Structure of the rhodococcus sp. usk13 darr-20 bp DNA complex
- Reference
- Schumacher MA, Lent N, Chen VB, Salinas R (2023): "Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding." Nat Commun, 14, 7239. doi: 10.1038/s41467-023-42823-0.
- Abstract
- The mycobacterial repressor, DarR, a TetR family regulator (TFR), was the first transcription regulator shown to bind c-di-AMP. However, the molecular basis for this interaction and the mechanism involved in DNA binding by DarR remain unknown. Here we describe DarR-c-di-AMP and DarR-DNA structures and complementary biochemical assays. The DarR-c-di-AMP structure reveals a unique effector binding site for a TFR, located between DarR dimer subunits. Strikingly, we show this motif also binds cAMP. The location of the adenine nucleotide binding site between subunits suggests this interaction may facilitate dimerization and hence DNA binding. Indeed, biochemical assays show cAMP enhances DarR DNA binding. Finally, DarR-DNA structures reveal a distinct TFR DNA-binding mechanism involving two interacting dimers on the DNA. Thus, the combined data unveil a newly described second messenger binding motif and DNA binding mode for this important family of regulators.