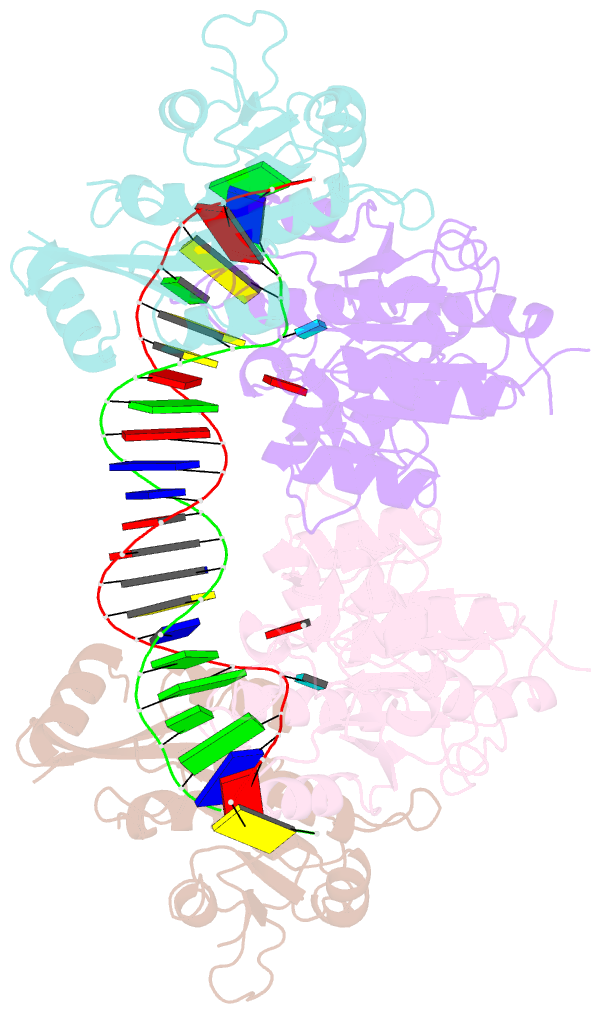
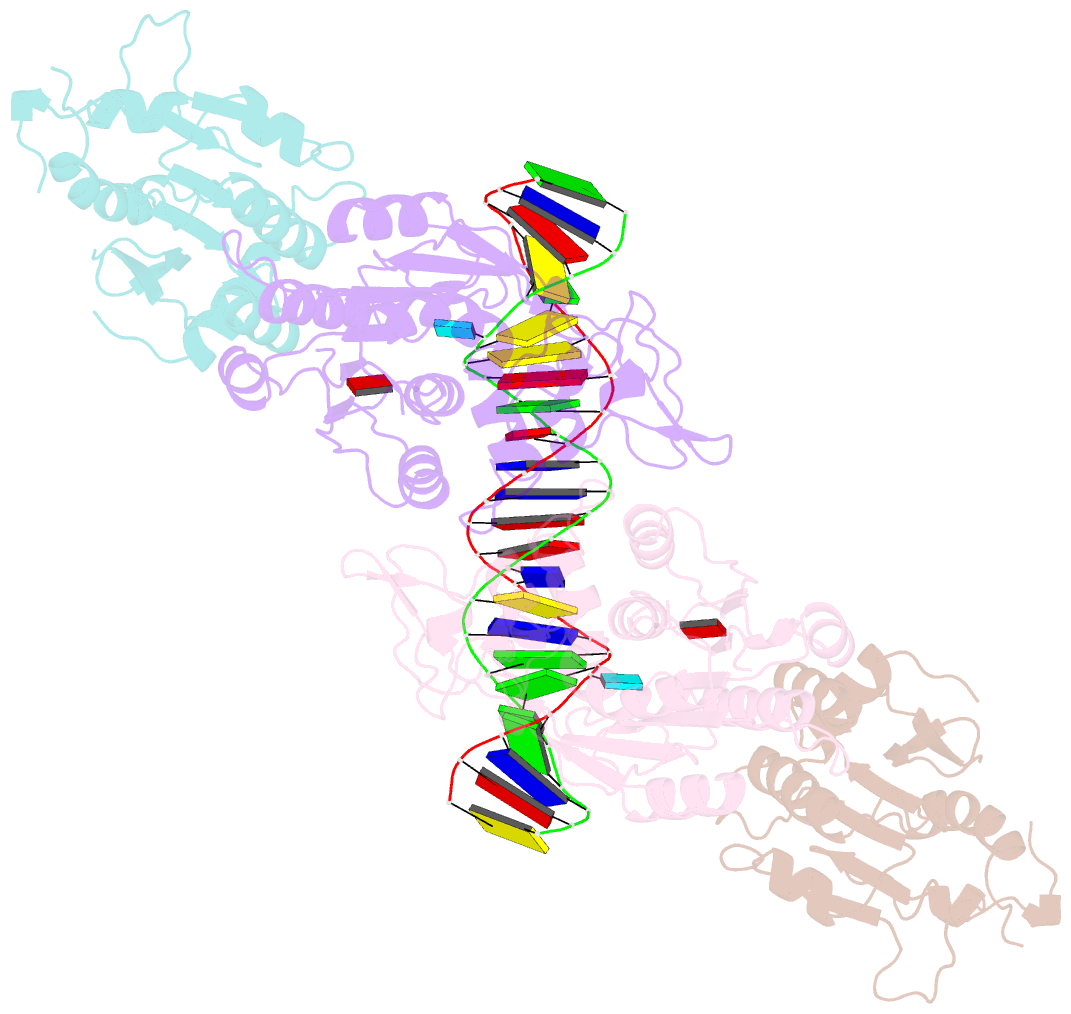
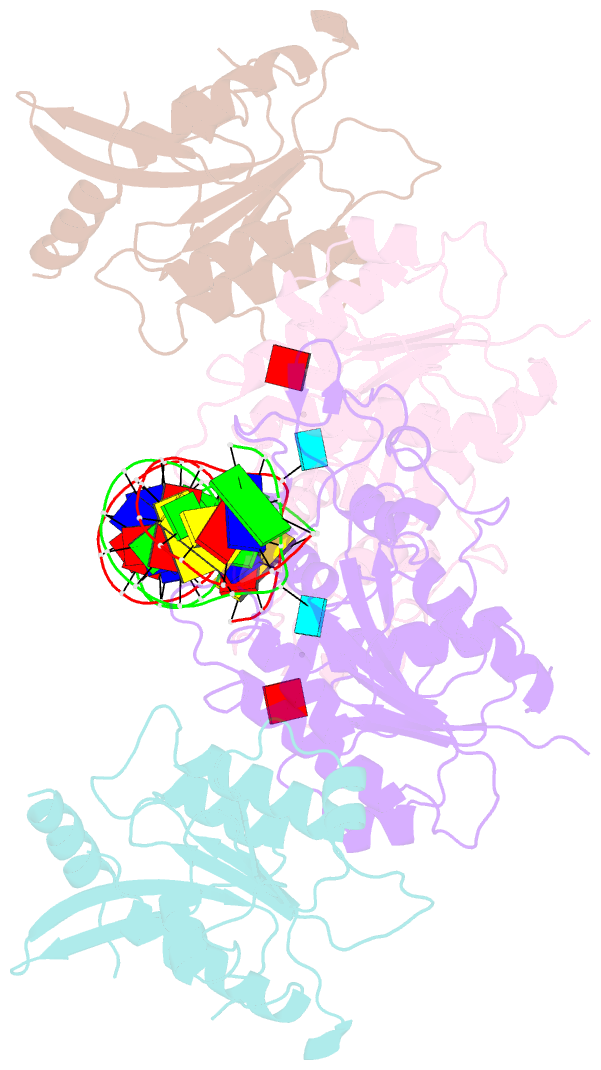
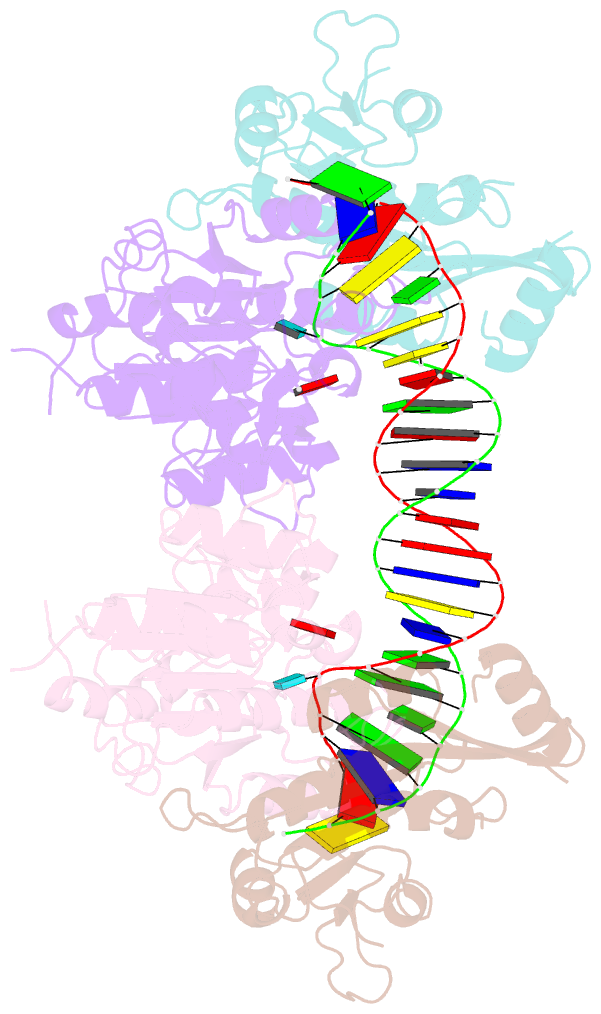
Summary information and primary citation
- PDB-id
- 8tci; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase
- Method
- X-ray (3.19 Å)
- Summary
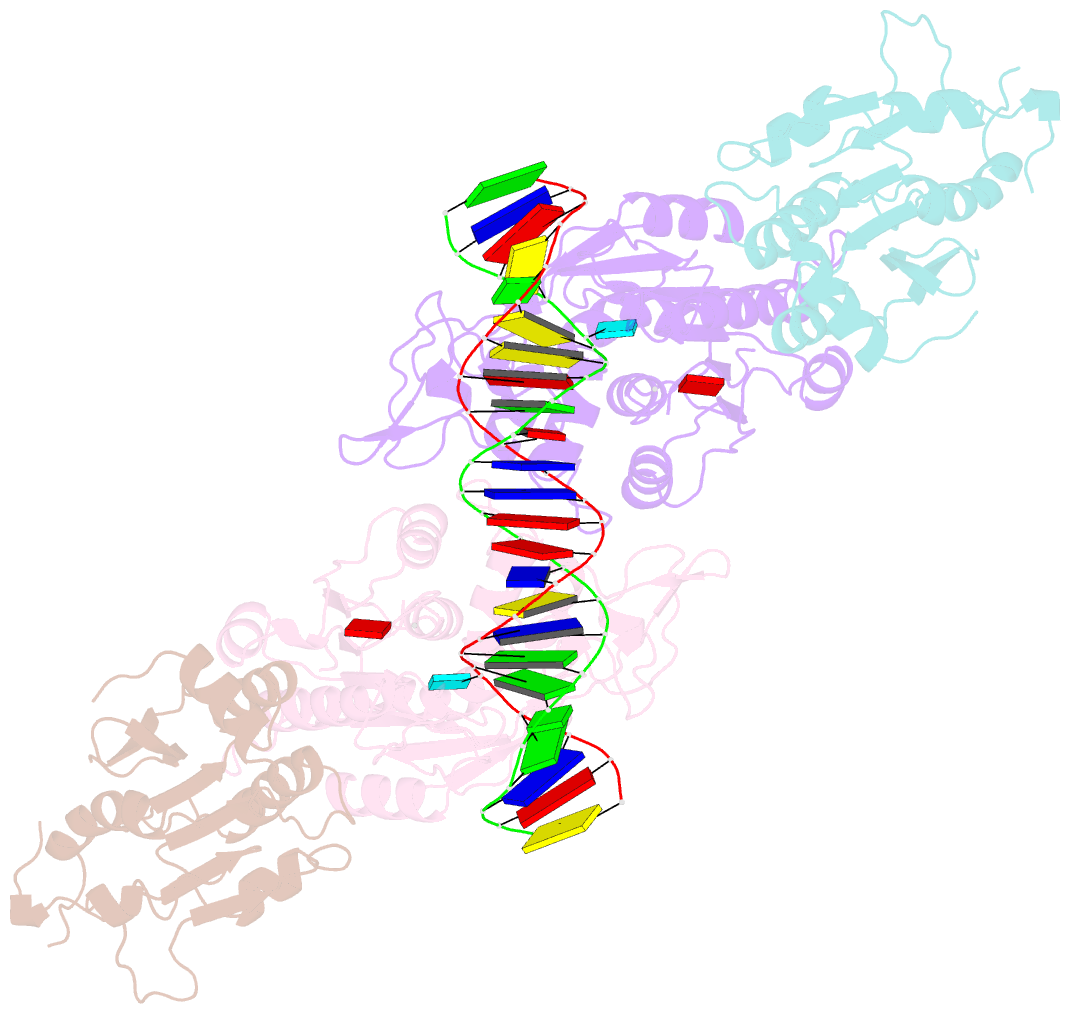
- Crystal structure of dnmt3c-dnmt3l in complex with cgg DNA
- Reference
- Khudaverdyan N, Lu J, Chen X, Herle G, Song J (2024): "The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation." J.Biol.Chem., 300, 107633. doi: 10.1016/j.jbc.2024.107633.
- Abstract
- DNA methylation is one of the major epigenetic mechanisms crucial for gene regulation and genome stability. De novo DNA methyltransferase DNMT3C is required for silencing evolutionarily young transposons during mice spermatogenesis. Mutation of DNMT3C led to a sterility phenotype that cannot be rescued by its homologues DNMT3A and DNMT3B. However, the structural basis of DNMT3C-mediated DNA methylation remains unknown. Here, we report the structure and mechanism of DNMT3C-mediated DNA methylation. The DNMT3C methyltransferase domain recognizes CpG-containing DNA in a manner similar to that of DNMT3A and DNMT3B, in line with their high sequence similarity. However, two evolutionary covariation sites, C543 and E590, diversify the substrate interaction among DNMT3C, DNMT3A and DNMT3B, resulting in distinct DNA methylation activity and specificity between DNMT3C, DNMT3A and DNMT3B in vitro. In addition, our combined structural and biochemical analysis reveals that the disease-causing rahu mutation of DNMT3C compromises its oligomerization and DNA-binding activities, providing an explanation for the loss of DNA methylation activity caused by this mutation. This study provides a mechanistic insight into DNMT3C-mediated DNA methylation that complements DNMT3A- and DNMT3B-mediated DNA methylation in mice, unravelling a regulatory mechanism by which evolutionary conservation and diversification fine-tunes the activity of de novo DNA methyltransferases.