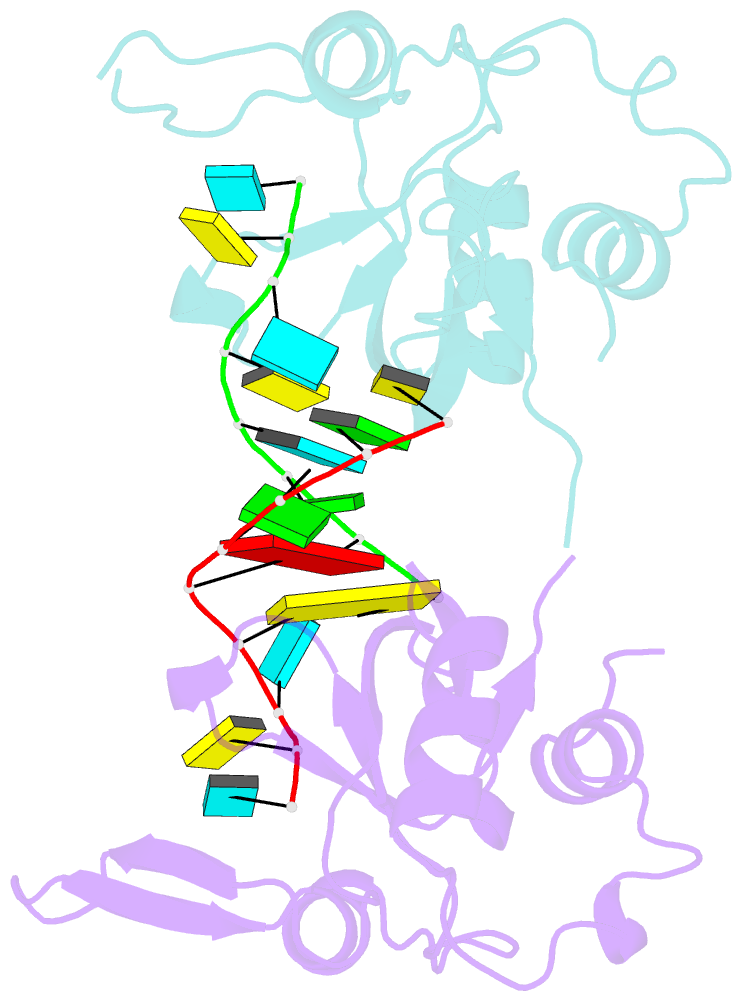
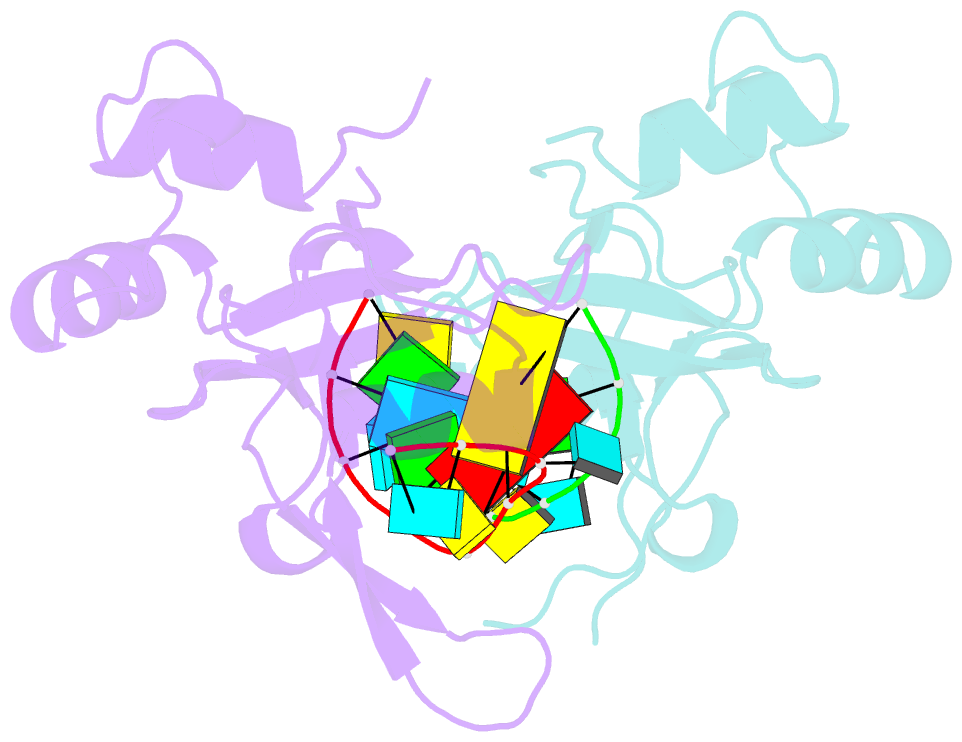
Summary information and primary citation
- PDB-id
- 8thq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.41 Å)
- Summary
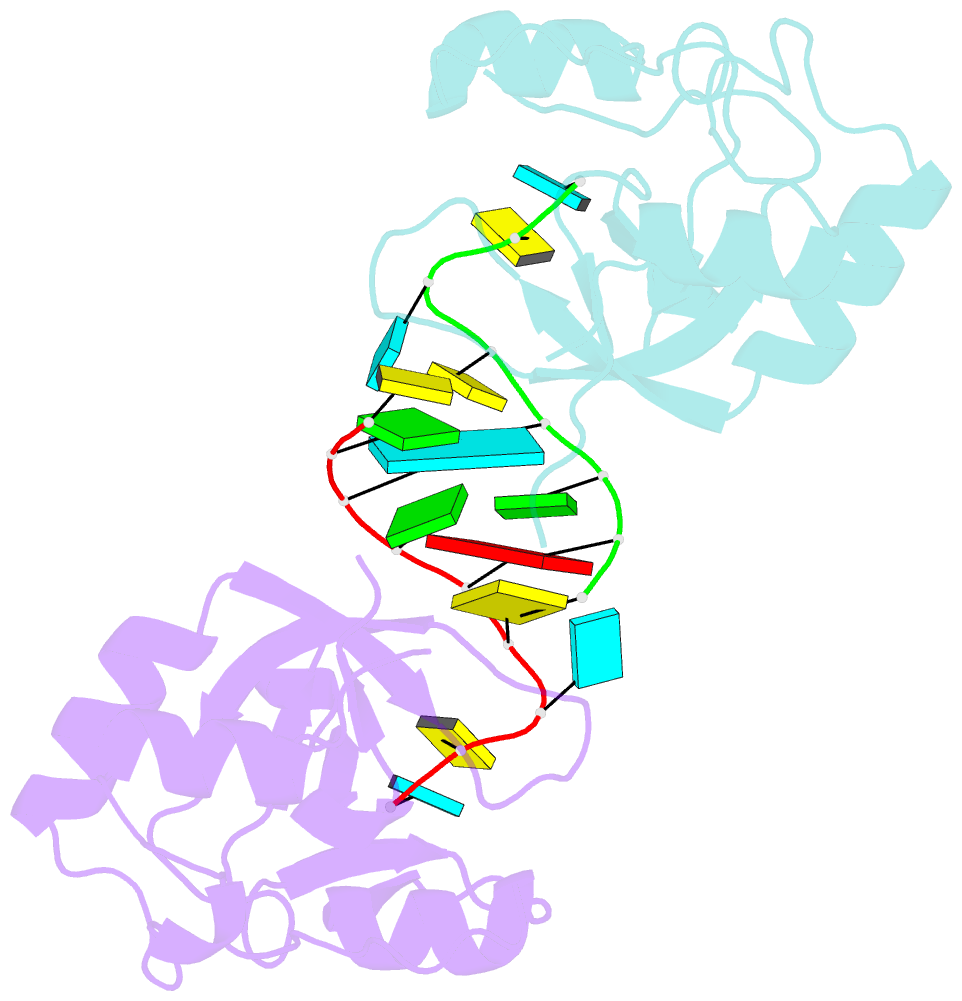
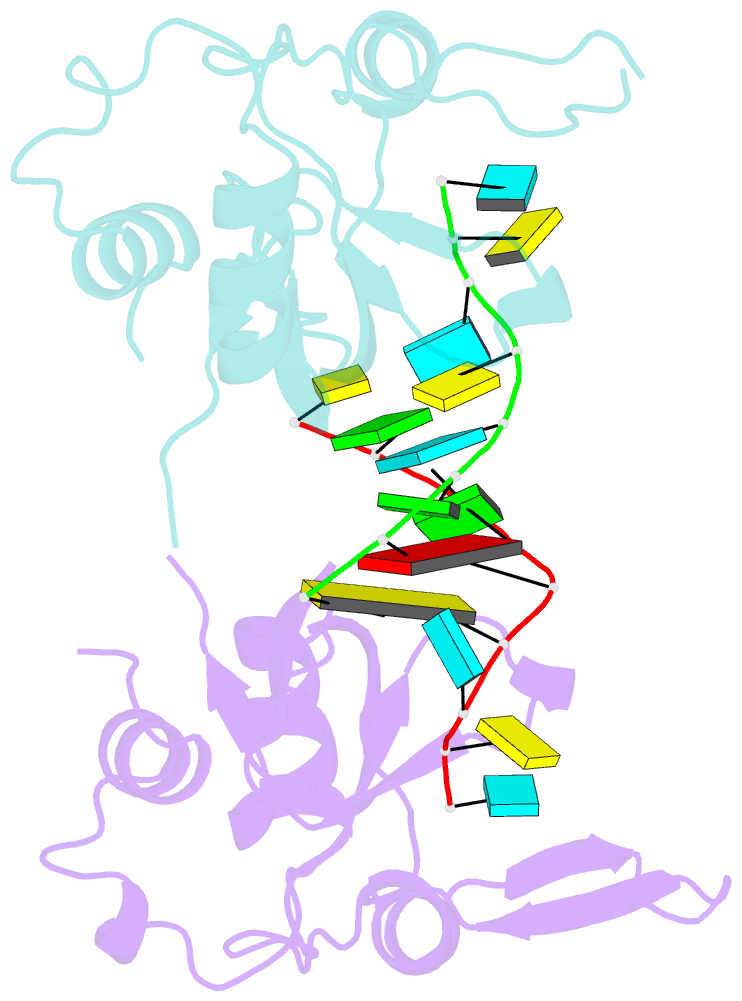
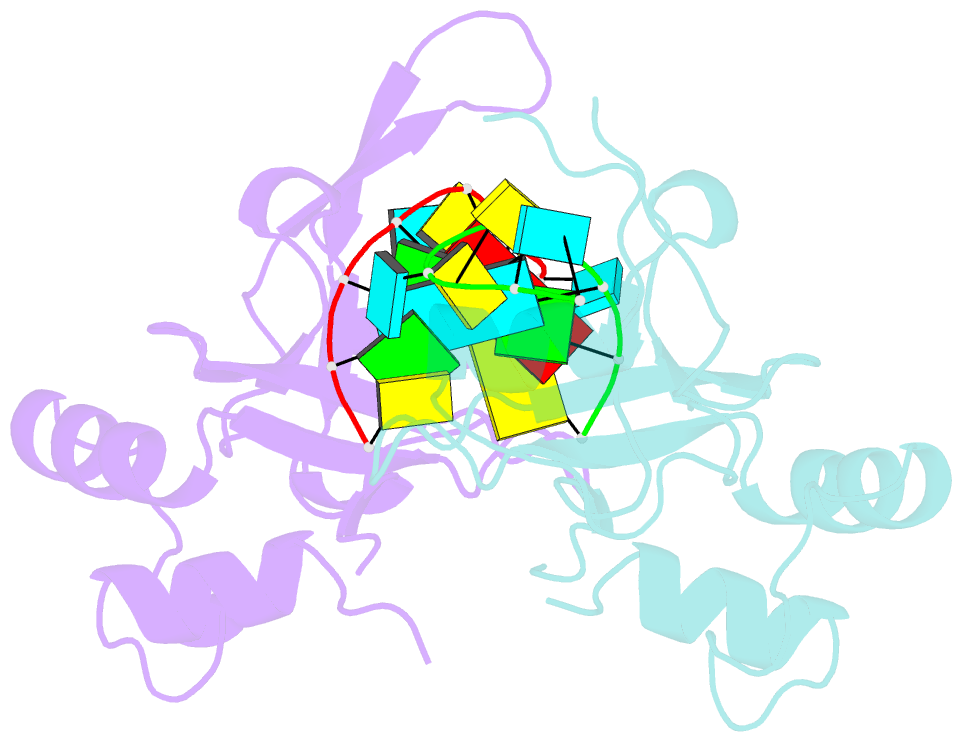
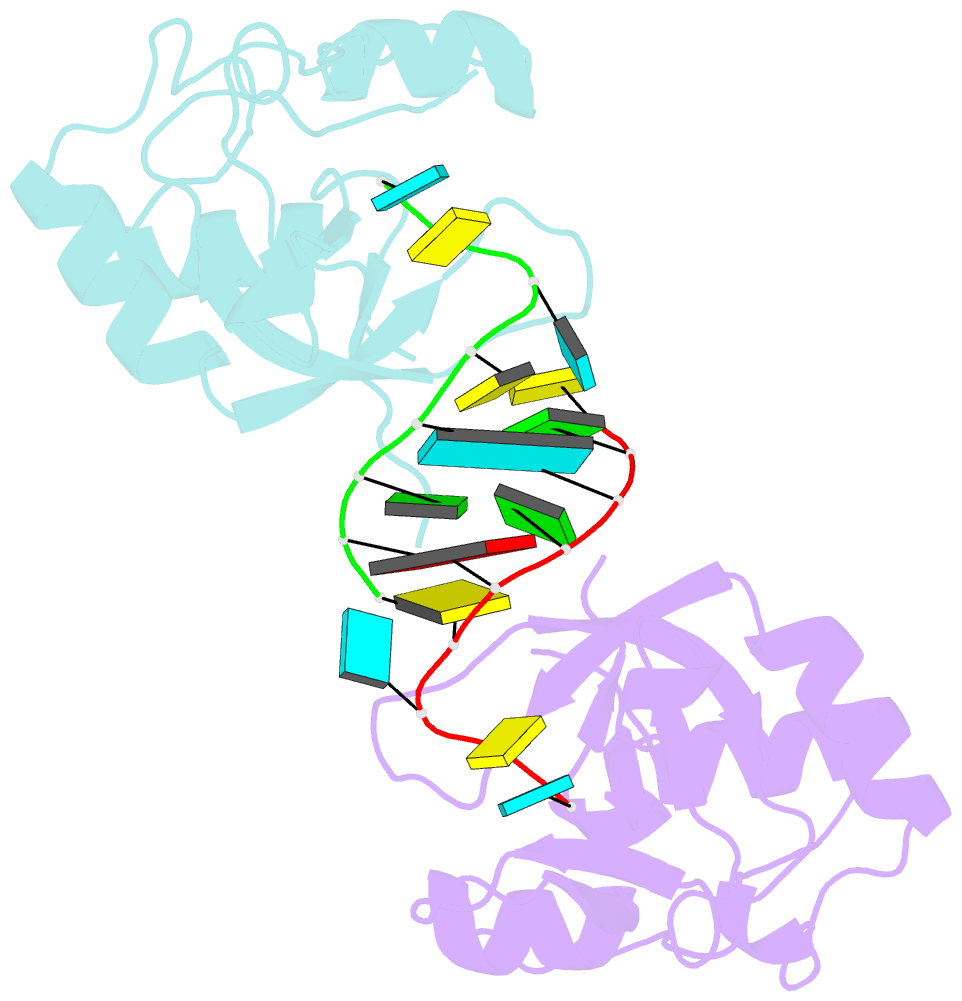
- Nonamer RNA bound to hago2-paz
- Reference
- Aluri KC, Datta D, Waldron S, Taneja N, Qin J, Donnelly DP, Theile CS, Guenther DC, Lei L, Harp JM, Pallan PS, Egli M, Zlatev I, Manoharan M (2024): "Single-Stranded Hairpin Loop RNAs (loopmeRNAs) Potently Induce Gene Silencing through the RNA Interference Pathway." J.Am.Chem.Soc. doi: 10.1021/jacs.4c07902.
- Abstract
- Synthetic small interfering RNAs conjugated to trivalent N-acetylgalactosamine (GalNAc) are clinically validated drugs for treatment of liver diseases. Incorporation of phosphorothioate linkages and ribose modifications are necessary for stability, potency, and duration of pharmacology. Although multiple alternative siRNA designs such as Dicer-substrate RNA, shRNA, and circular RNA have been evaluated in vitro and in preclinical studies with some success, clinical applications of these designs are limited as it is difficult to incorporate chemical modifications in these designs. An alternative siRNA design that can incorporate chemical modifications through straightforward synthesis without compromising potency will significantly advance the field. Here, we report a facile synthesis of GalNAc ligand-containing single-stranded loop hairpin RNAs (loopmeRNAs) with clinically relevant chemical modifications. We evaluated the efficiency of novel loopmeRNA designs in vivo and correlated their structure-activity relationship with the support of in vitro metabolism data. Sequences and chemical modifications in the loop region of the loopmeRNA design were optimized for maximal potency. Our studies demonstrate that loopmeRNAs can efficiently silence expression of target genes with comparable efficacy to conventional double-stranded siRNAs but reduced environmental and regulatory burdens.