Summary information and primary citation
- PDB-id
- 8tl0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system
- Method
- cryo-EM (3.1 Å)
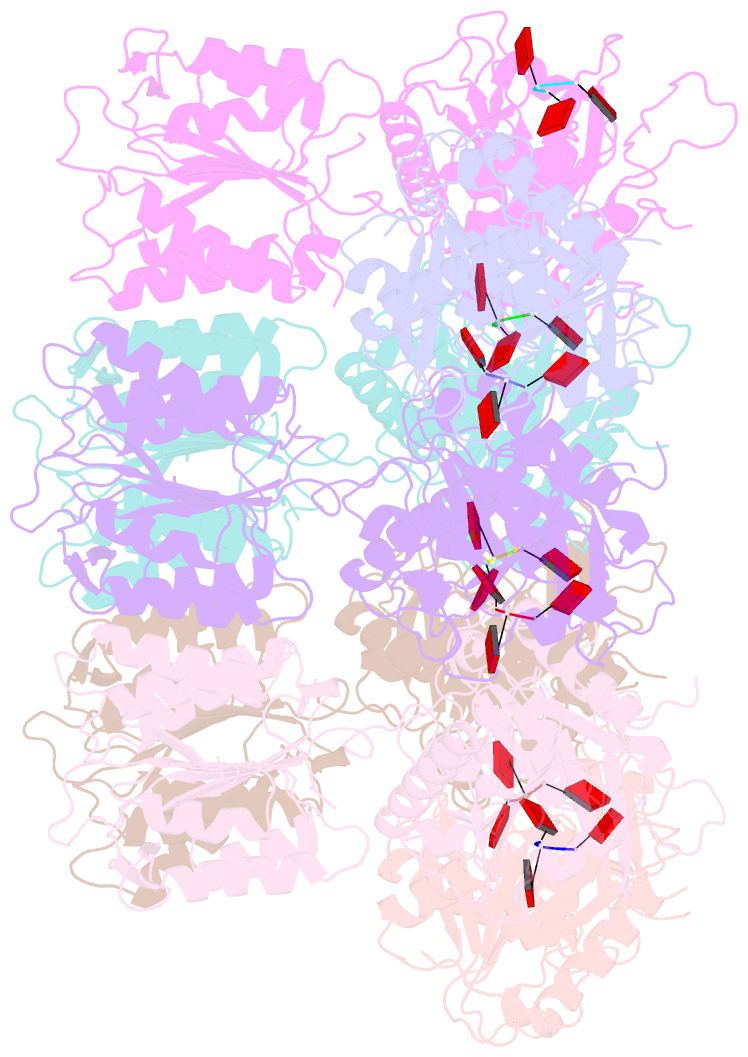
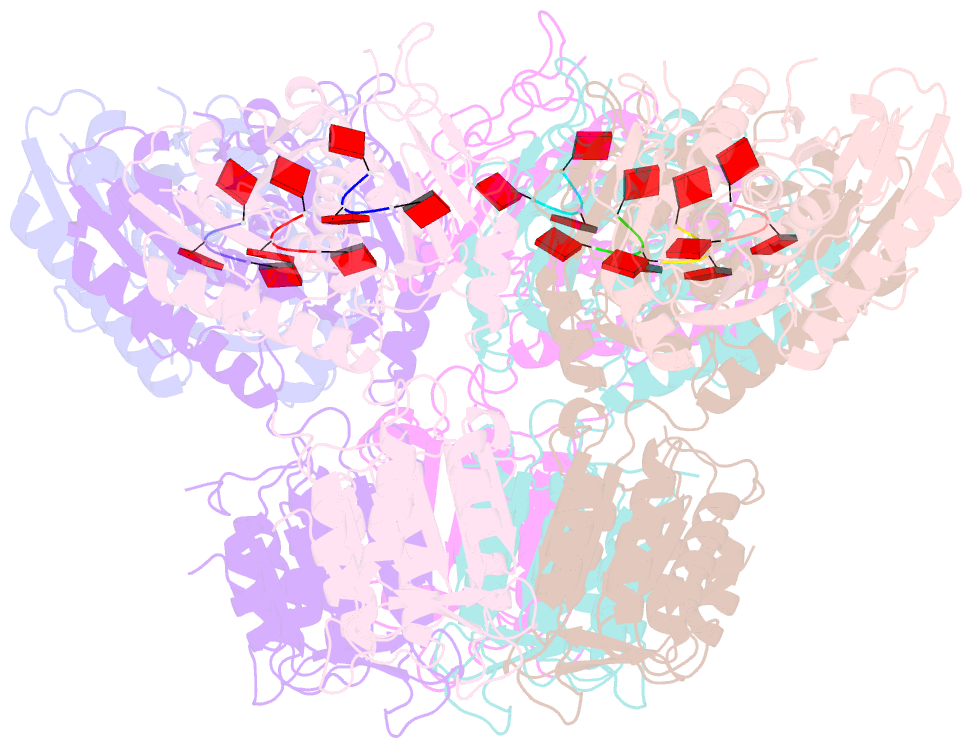
- Summary
- Structure of activated saved-chat filament
- Reference
- Steens JA, Bravo JPK, Salazar CRP, Yildiz C, Amieiro AM, Kostlbacher S, Prinsen SHP, Andres AS, Patinios C, Bardis A, Barendregt A, Scheltema RA, Ettema TJG, van der Oost J, Taylor DW, Staals RHJ (2024): "Type III-B CRISPR-Cas cascade of proteolytic cleavages." Science, 383, 512-519. doi: 10.1126/science.adk0378.
- Abstract
- The generation of cyclic oligoadenylates and subsequent allosteric activation of proteins that carry sensory domains is a distinctive feature of type III CRISPR-Cas systems. In this work, we characterize a set of associated genes of a type III-B system from Haliangium ochraceum that contains two caspase-like proteases, SAVED-CHAT and PCaspase (prokaryotic caspase), co-opted from a cyclic oligonucleotide-based antiphage signaling system (CBASS). Cyclic tri-adenosine monophosphate (AMP)-induced oligomerization of SAVED-CHAT activates proteolytic activity of the CHAT domains, which specifically cleave and activate PCaspase. Subsequently, activated PCaspase cleaves a multitude of proteins, which results in a strong interference phenotype in vivo in Escherichia coli. Taken together, our findings reveal how a CRISPR-Cas-based detection of a target RNA triggers a cascade of caspase-associated proteolytic activities.