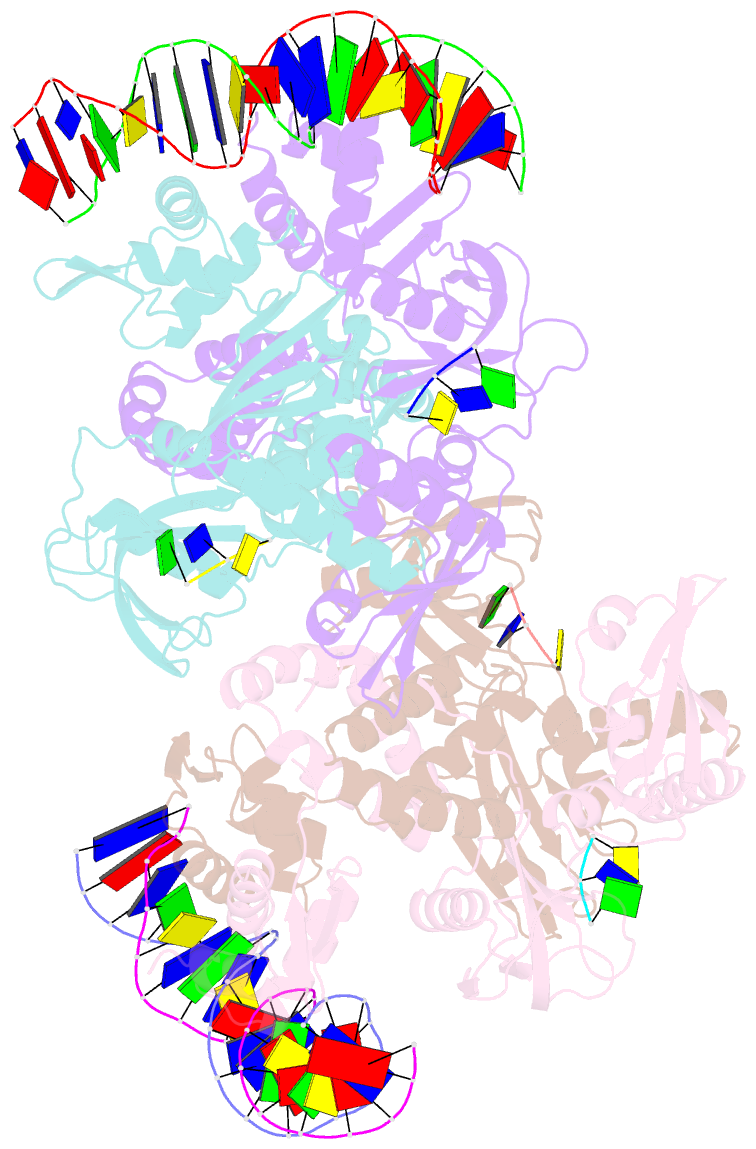
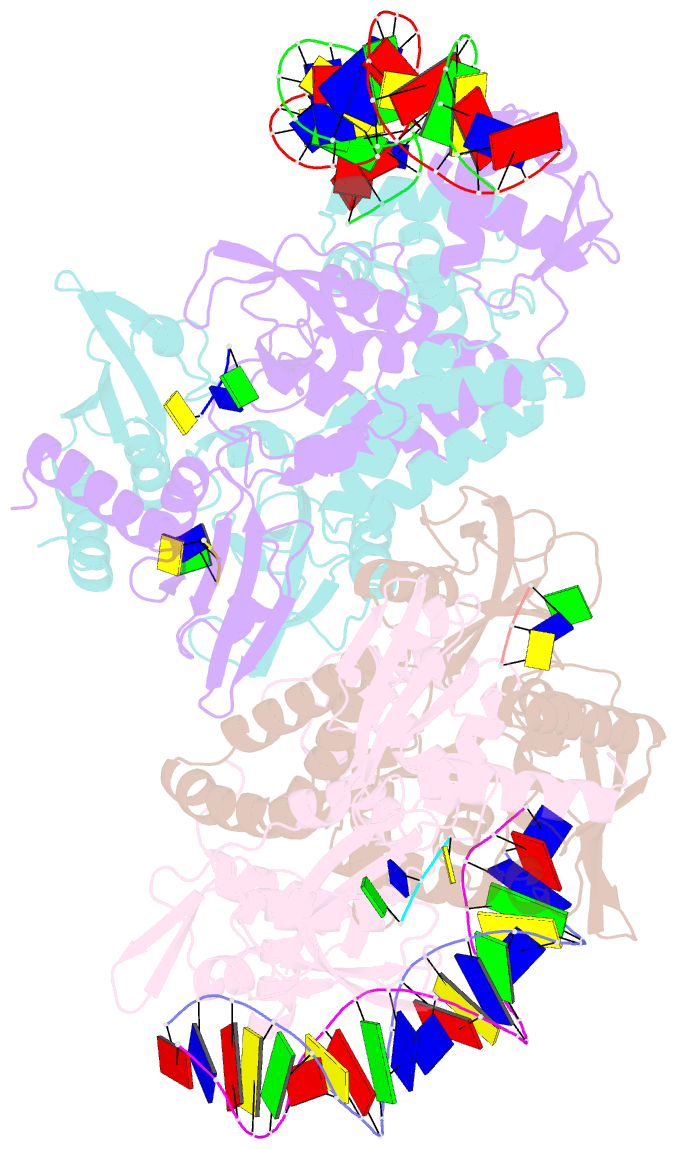
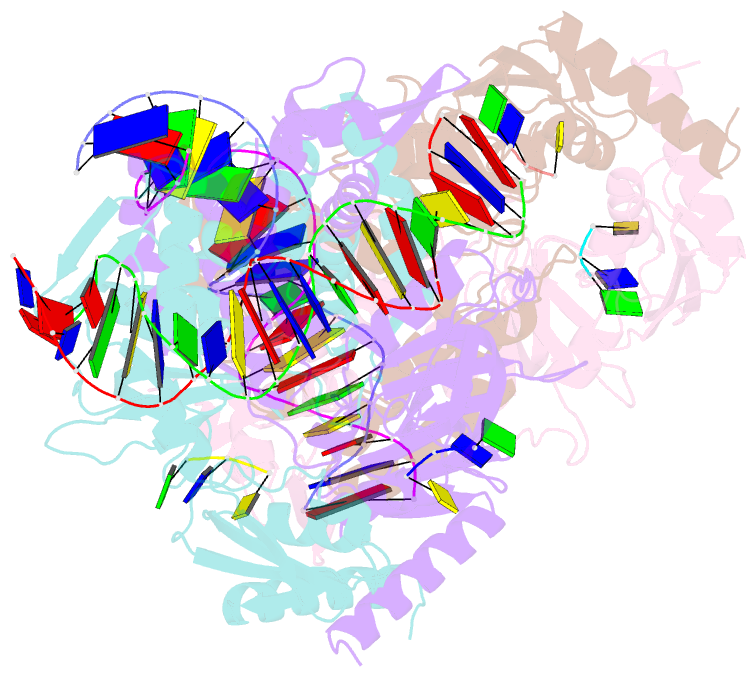
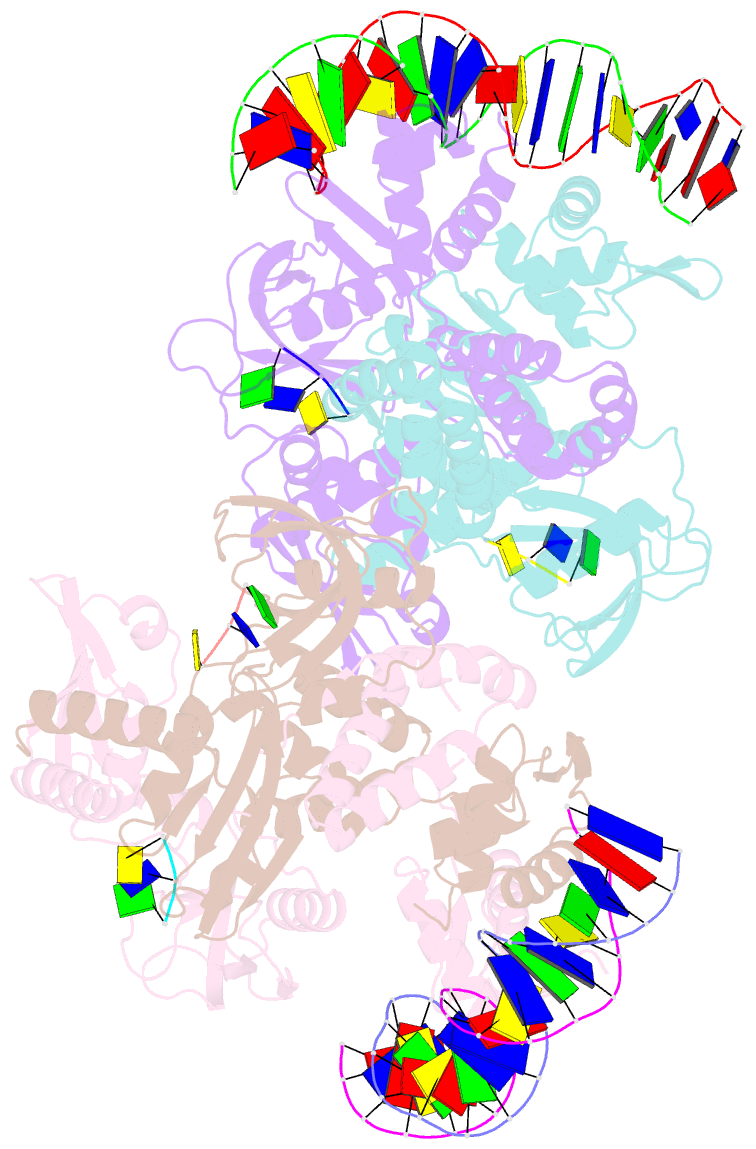
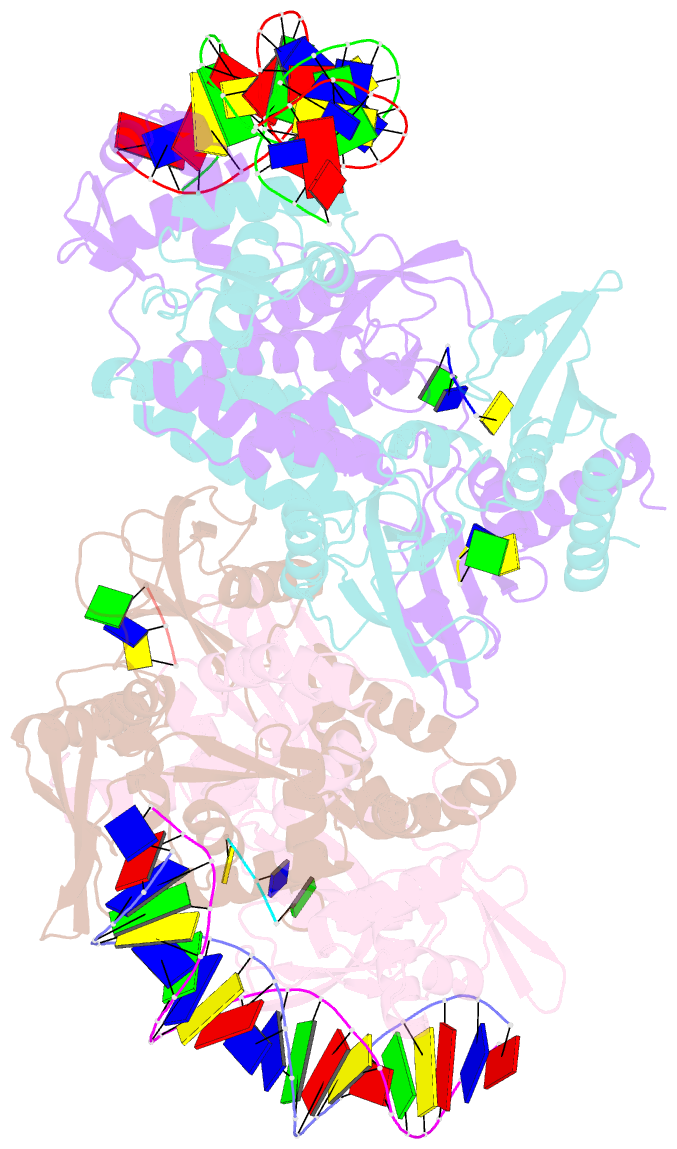
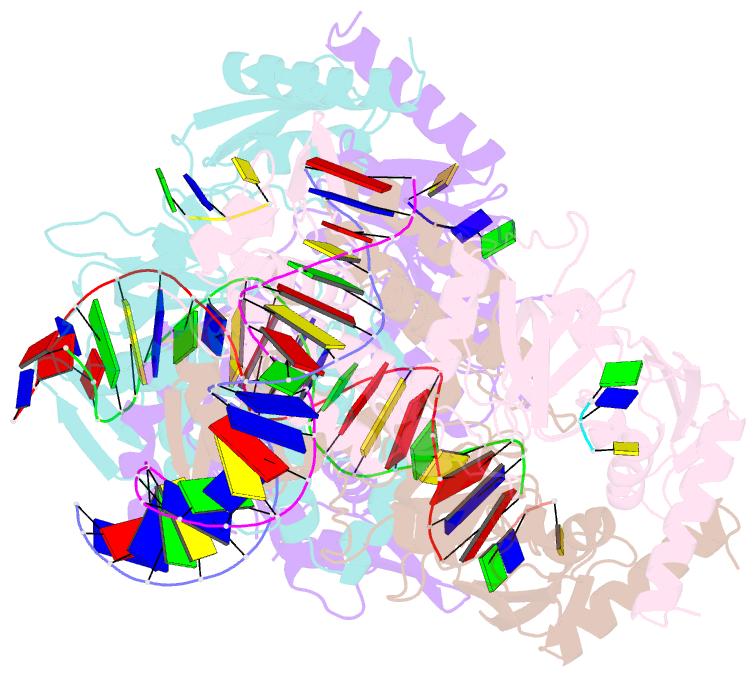
Summary information and primary citation
- PDB-id
- 8tp8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.74 Å)
- Summary
- Structure of the c. crescentus wyl-activator, drid, bound to ssDNA and cognate DNA
- Reference
- Schumacher MA, Cannistraci E, Salinas R, Lloyd D, Messner E, Gozzi K (2024): "Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site." Nucleic Acids Res., 52, 1435-1449. doi: 10.1093/nar/gkad1198.
- Abstract
- Transcription regulators play central roles in orchestrating responses to changing environmental conditions. Recently the Caulobacter crescentus transcription activator DriD, which belongs to the newly defined WYL-domain family, was shown to regulate DNA damage responses independent of the canonical SOS pathway. However, the molecular mechanisms by which DriD and other WYL-regulators sense environmental signals and recognize DNA are not well understood. We showed DriD DNA-binding is triggered by its interaction with ssDNA, which is produced during DNA damage. Here we describe the structure of the full-length C. crescentus DriD bound to both target DNA and effector ssDNA. DriD consists of an N-terminal winged-HTH (wHTH) domain, linker region, three-helix bundle, WYL-domain and C-terminal WCX-dimer domain. Strikingly, DriD binds DNA using a novel, asymmetric DNA-binding mechanism that results from different conformations adopted by the linker. Although the linker does not touch DNA, our data show that contacts it makes with the wHTH are key for specific DNA binding. The structure indicates how ssDNA-effector binding to the WYL-domain impacts wHTH DNA binding. In conclusion, we present the first structure of a WYL-activator bound to both effector and target DNA. The structure unveils a unique, asymmetric DNA binding mode that is likely conserved among WYL-activators.