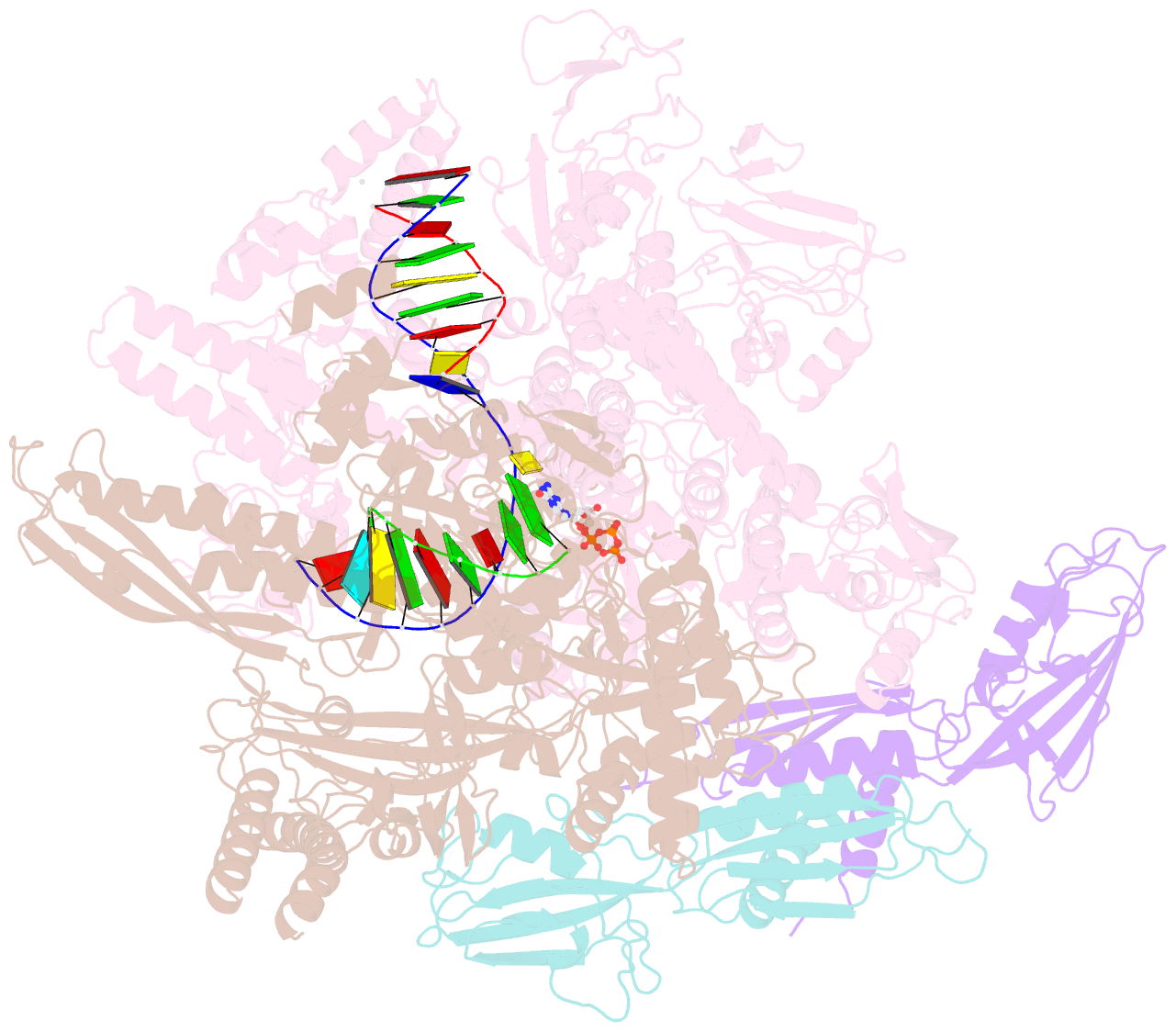
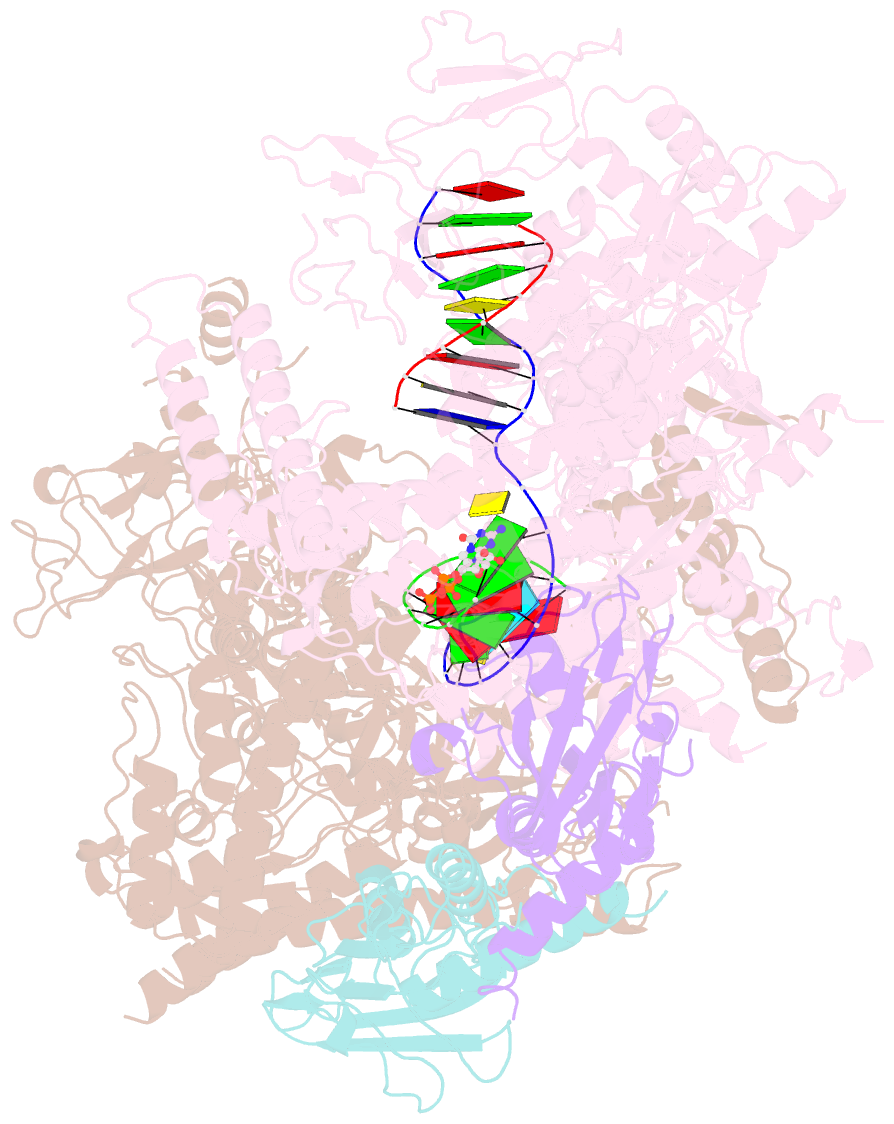
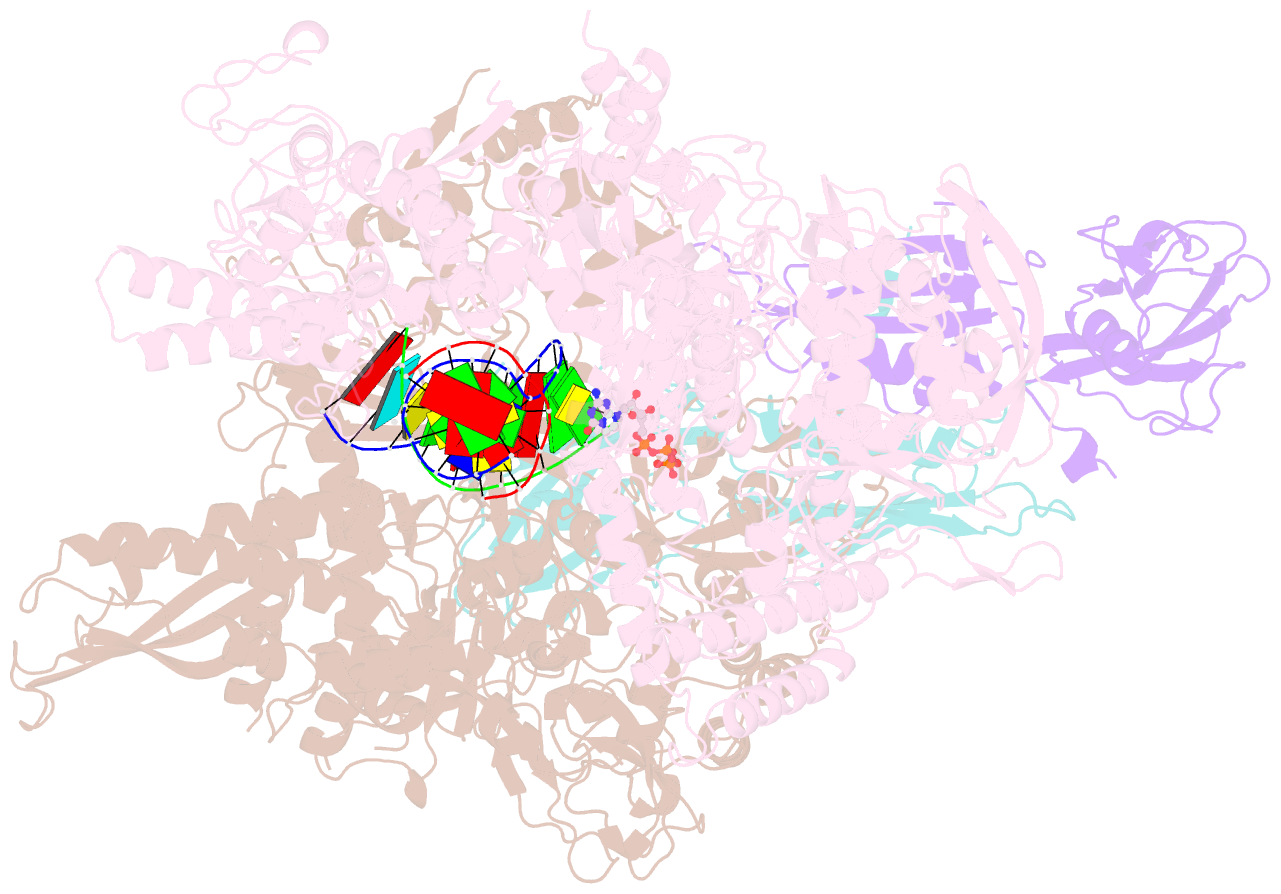
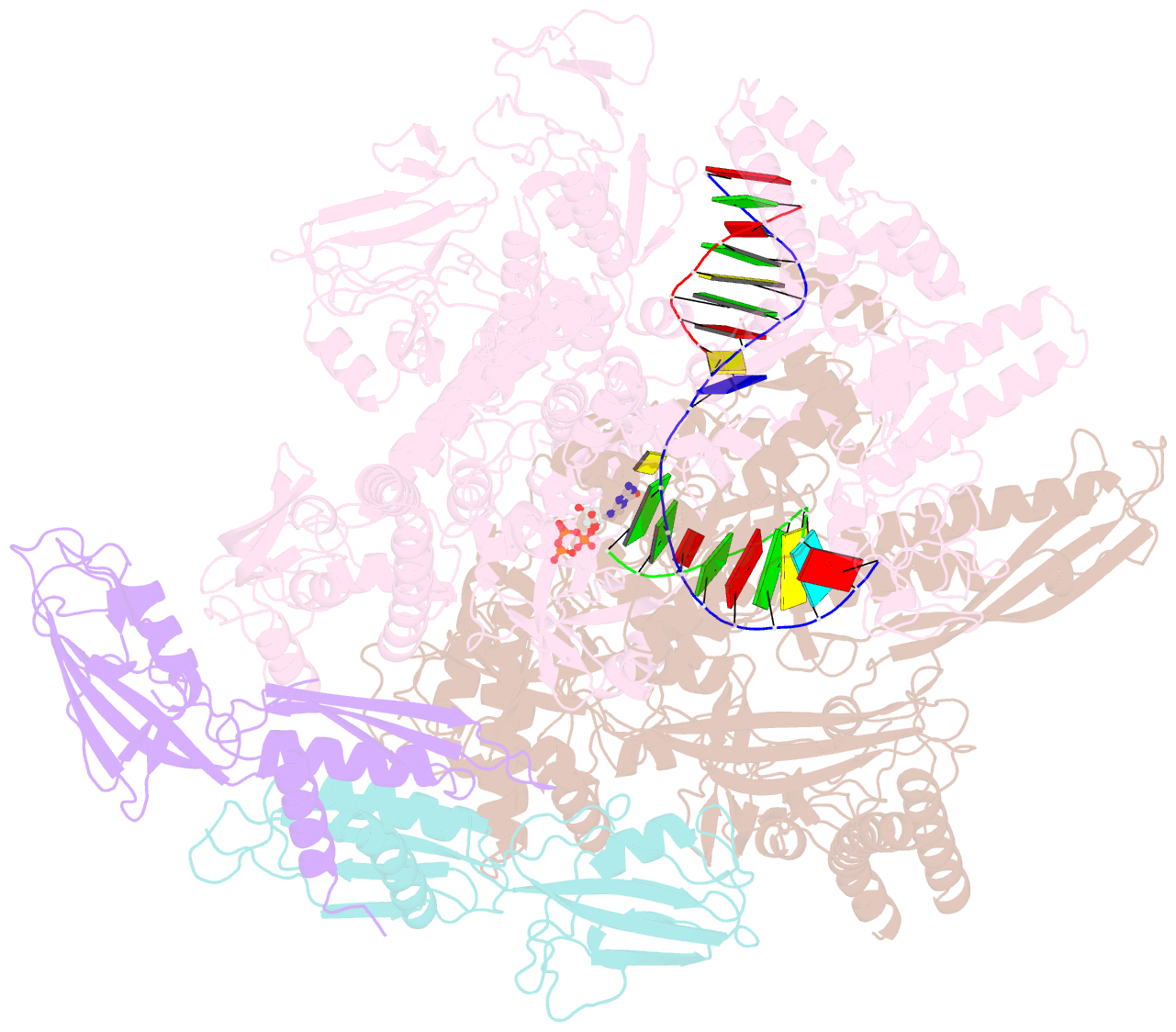
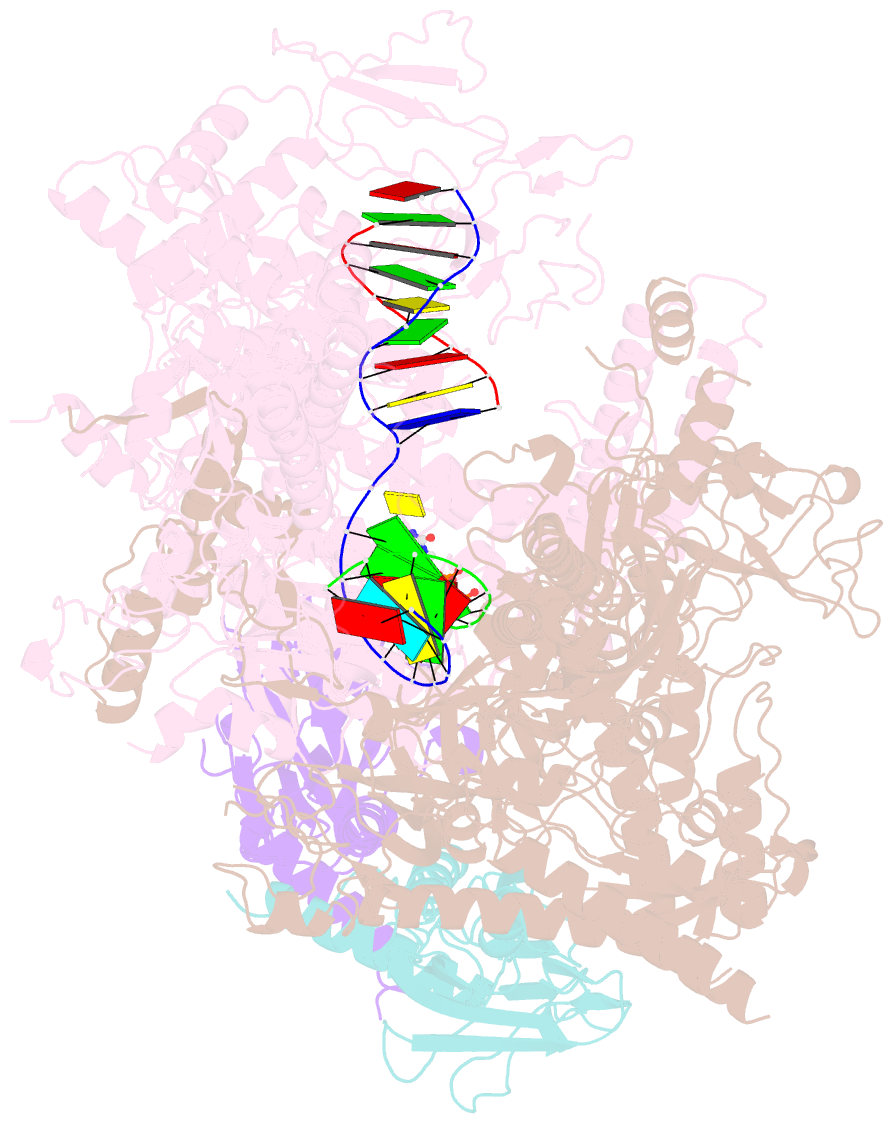
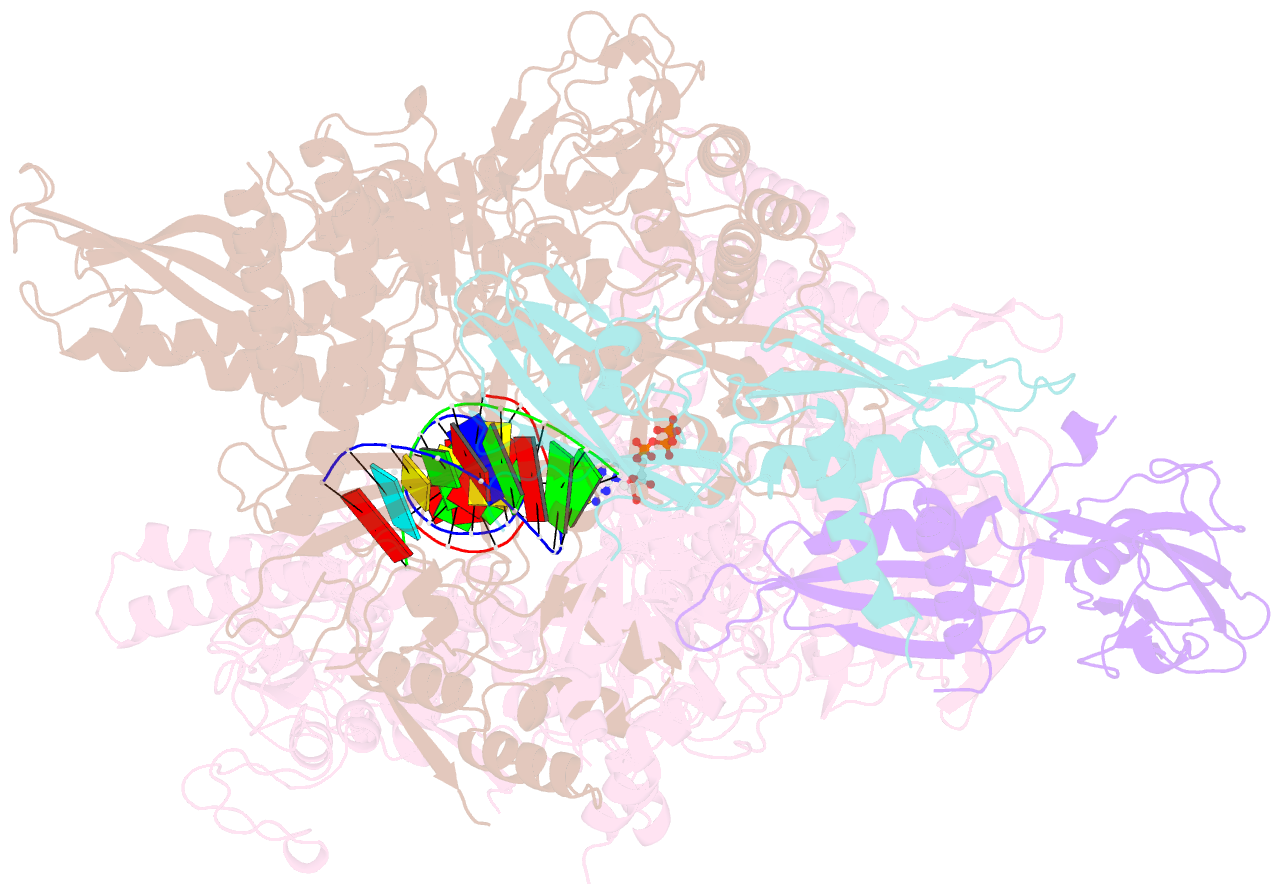
Summary information and primary citation
- PDB-id
- 8txo; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.1 Å)
- Summary
- E. coli DNA-directed RNA polymerase transcription elongation complex bound to the unnatural dz-ptp base pair in the active site
- Reference
- Aiyer S, Baldwin PR, Tan SM, Shan Z, Oh J, Mehrani A, Bowman ME, Louie G, Passos DO, Dordevic-Marquardt S, Mietzsch M, Hull JA, Hoshika S, Barad BA, Grotjahn DA, McKenna R, Agbandje-McKenna M, Benner SA, Noel JAP, Wang D, Tan YZ, Lyumkis D (2024): "Overcoming resolution attenuation during tilted cryo-EM data collection." Nat Commun, 15, 389. doi: 10.1038/s41467-023-44555-7.
- Abstract
- Structural biology efforts using cryogenic electron microscopy are frequently stifled by specimens adopting "preferred orientations" on grids, leading to anisotropic map resolution and impeding structure determination. Tilting the specimen stage during data collection is a generalizable solution but has historically led to substantial resolution attenuation. Here, we develop updated data collection and image processing workflows and demonstrate, using multiple specimens, that resolution attenuation is negligible or significantly reduced across tilt angles. Reconstructions with and without the stage tilted as high as 60° are virtually indistinguishable. These strategies allowed the reconstruction to 3 Å resolution of a bacterial RNA polymerase with preferred orientation, containing an unnatural nucleotide for studying novel base pair recognition. Furthermore, we present a quantitative framework that allows cryo-EM practitioners to define an optimal tilt angle during data acquisition. These results reinforce the utility of employing stage tilt for data collection and provide quantitative metrics to obtain isotropic maps.