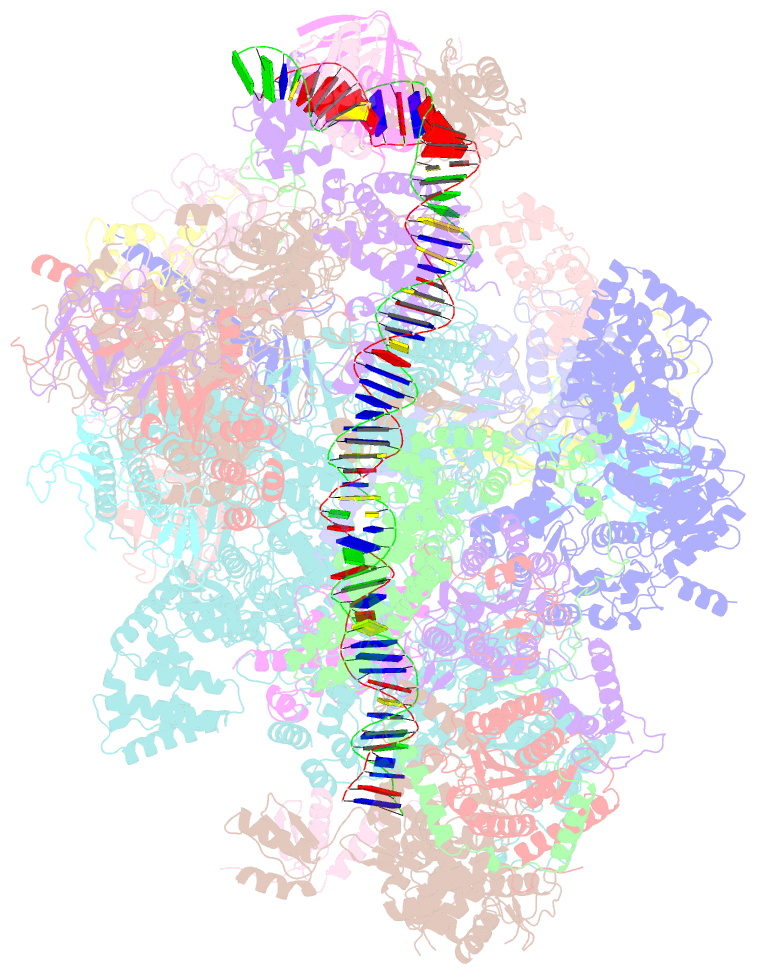
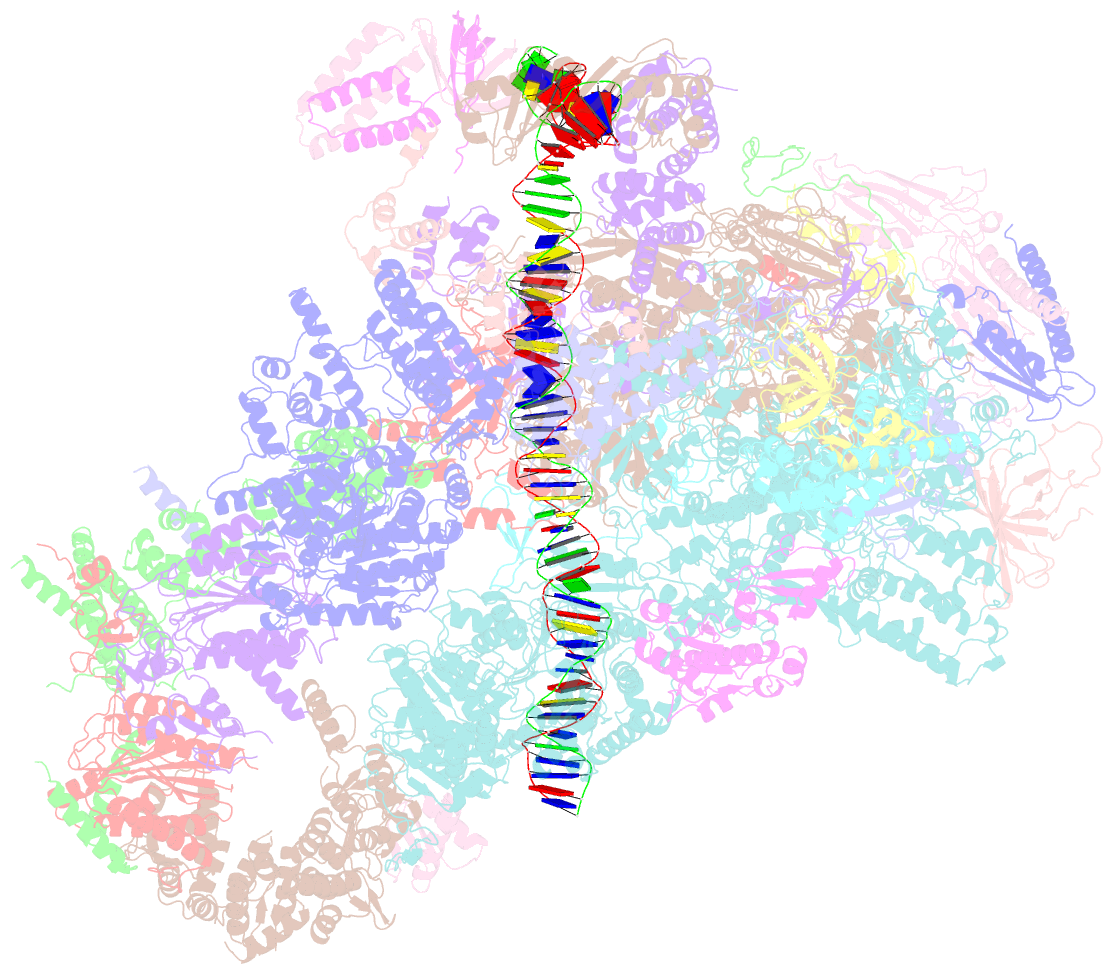
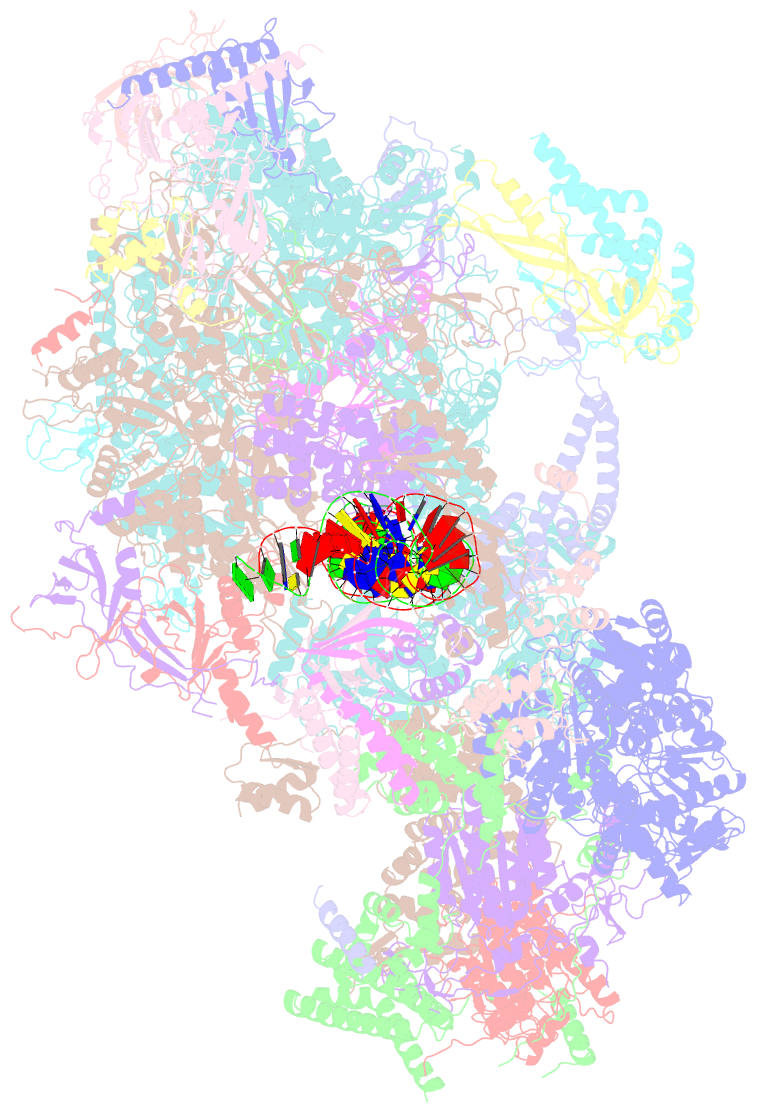
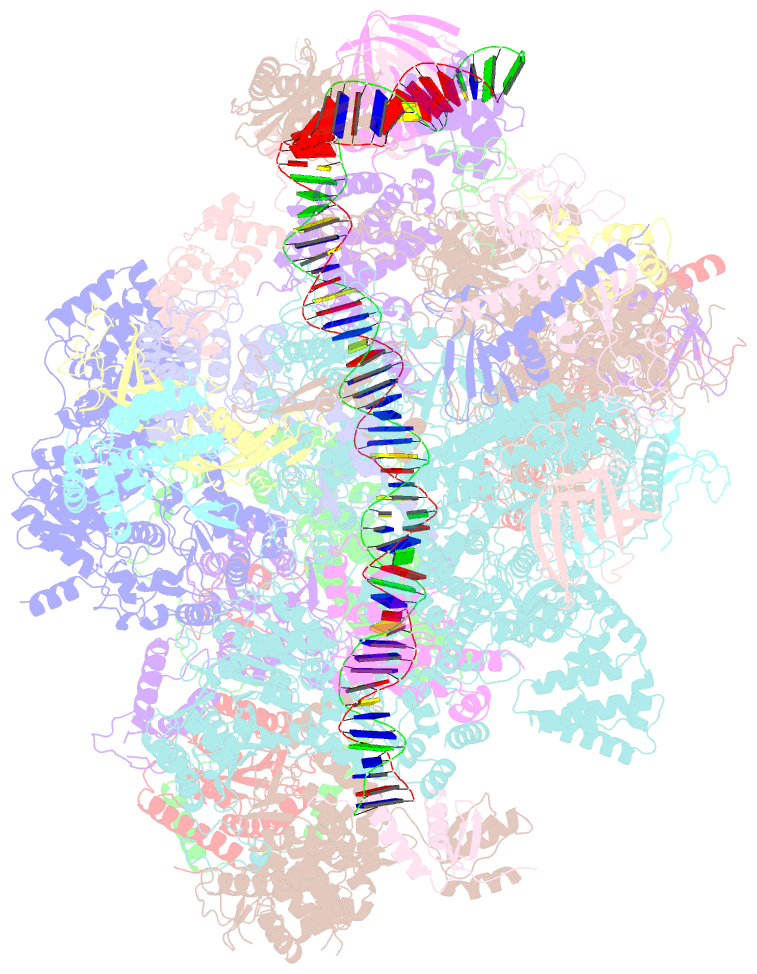
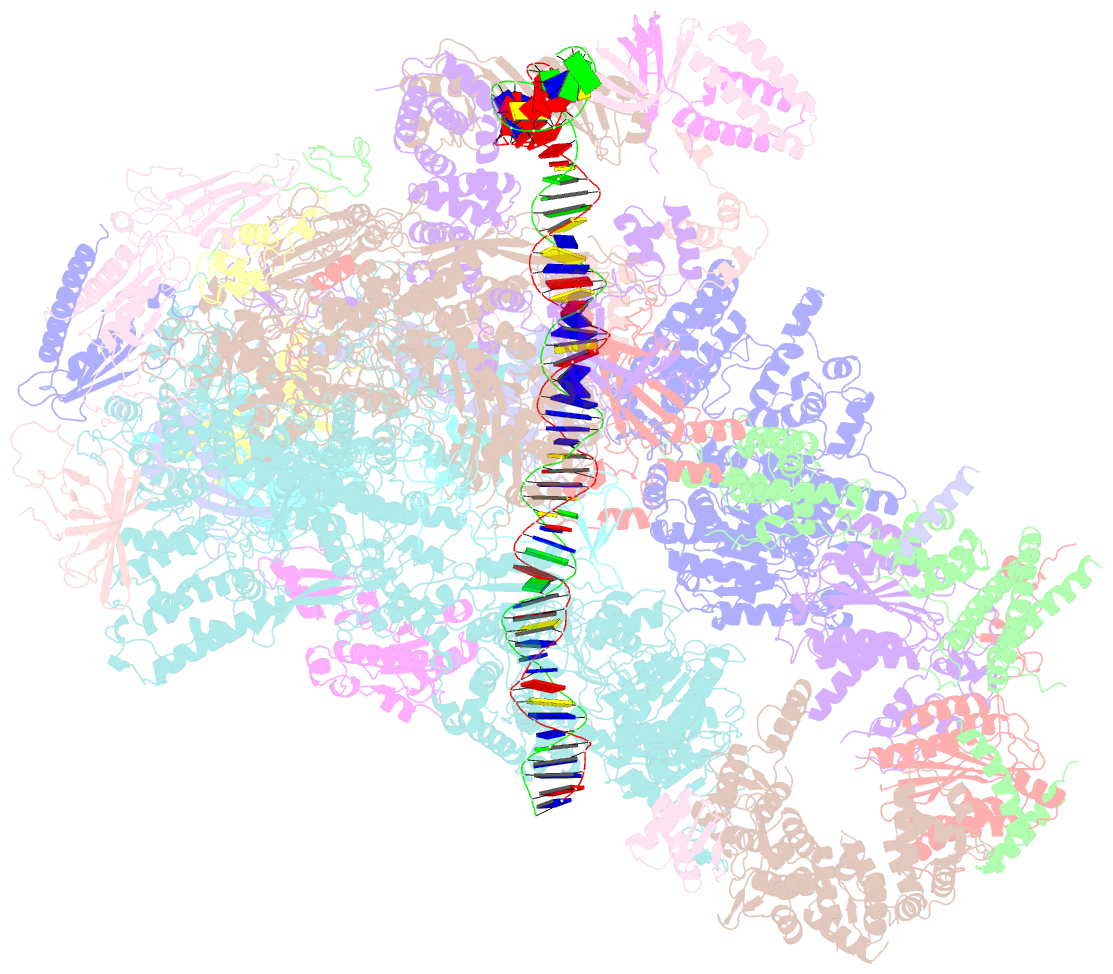
Summary information and primary citation
- PDB-id
- 8uoq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.8 Å)
- Summary
- Composite map of pic_delta_tfiik form2
- Reference
- Yang C, Basnet P, Sharmin S, Shen H, Kaplan CD, Murakami K (2024): "Transcription start site scanning requires the fungi-specific hydrophobic loop of Tfb3." Nucleic Acids Res., 52, 11602-11611. doi: 10.1093/nar/gkae805.
- Abstract
- RNA polymerase II (pol II) initiates transcription from transcription start sites (TSSs) located ∼30-35 bp downstream of the TATA box in metazoans, whereas in the yeast Saccharomyces cerevisiae, pol II scans further downstream TSSs located ∼40-120 bp downstream of the TATA box. Previously, we found that removal of the kinase module TFIIK (Kin28-Ccl1-Tfb3) from TFIIH shifts the TSS in a yeast in vitro system upstream to the location observed in metazoans and that addition of recombinant Tfb3 back to TFIIH-ΔTFIIK restores the downstream TSS usage. Here, we report that this biochemical activity of yeast TFIIK in TSS scanning is attributable to the Tfb3 RING domain at the interface with pol II in the pre-initiation complex (PIC): especially, swapping Tfb3 Pro51-a residue conserved among all fungi-with Ala or Ser as in MAT1, the metazoan homolog of Tfb3, confers an upstream TSS shift in vitro in a similar manner to the removal of TFIIK. Yeast genetic analysis suggests that both Pro51 and Arg64 of Tfb3 are required to maintain the stability of the Tfb3-pol II interface in the PIC. Cryo-electron microscopy analysis of a yeast PIC lacking TFIIK reveals considerable variability in the orientation of TFIIH, which impairs TSS scanning after promoter opening.