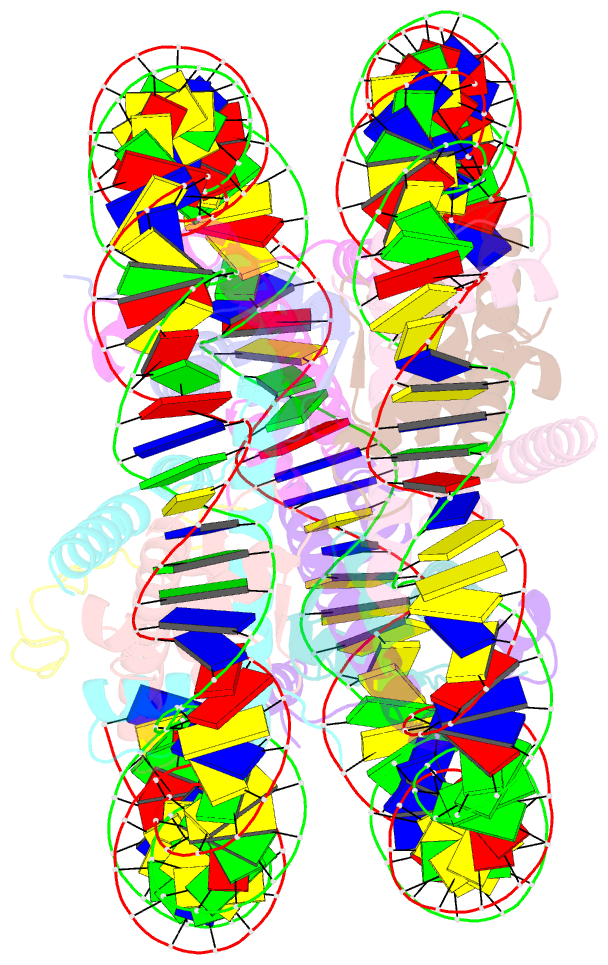
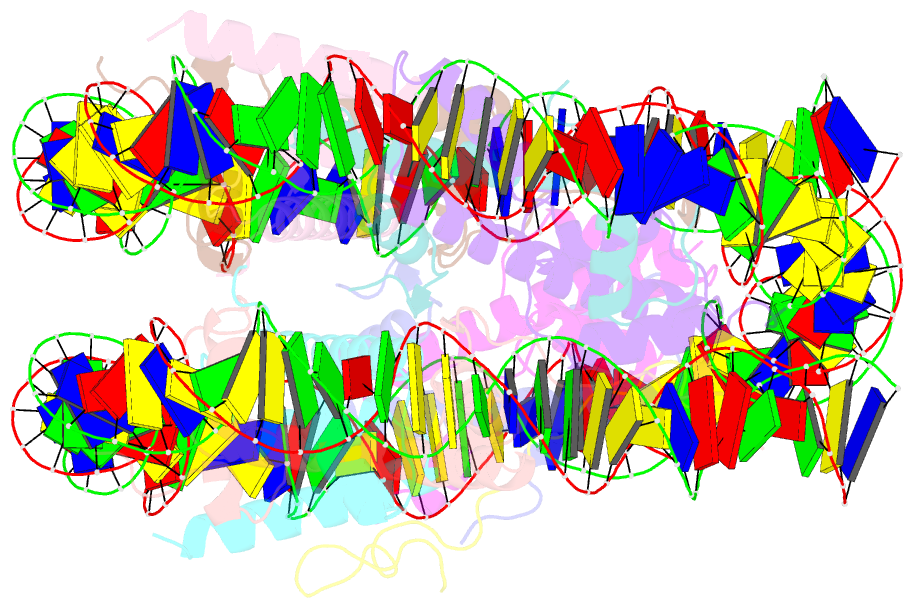
Summary information and primary citation
- PDB-id
- 8uw1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- gene regulation
- Method
- cryo-EM (2.88 Å)
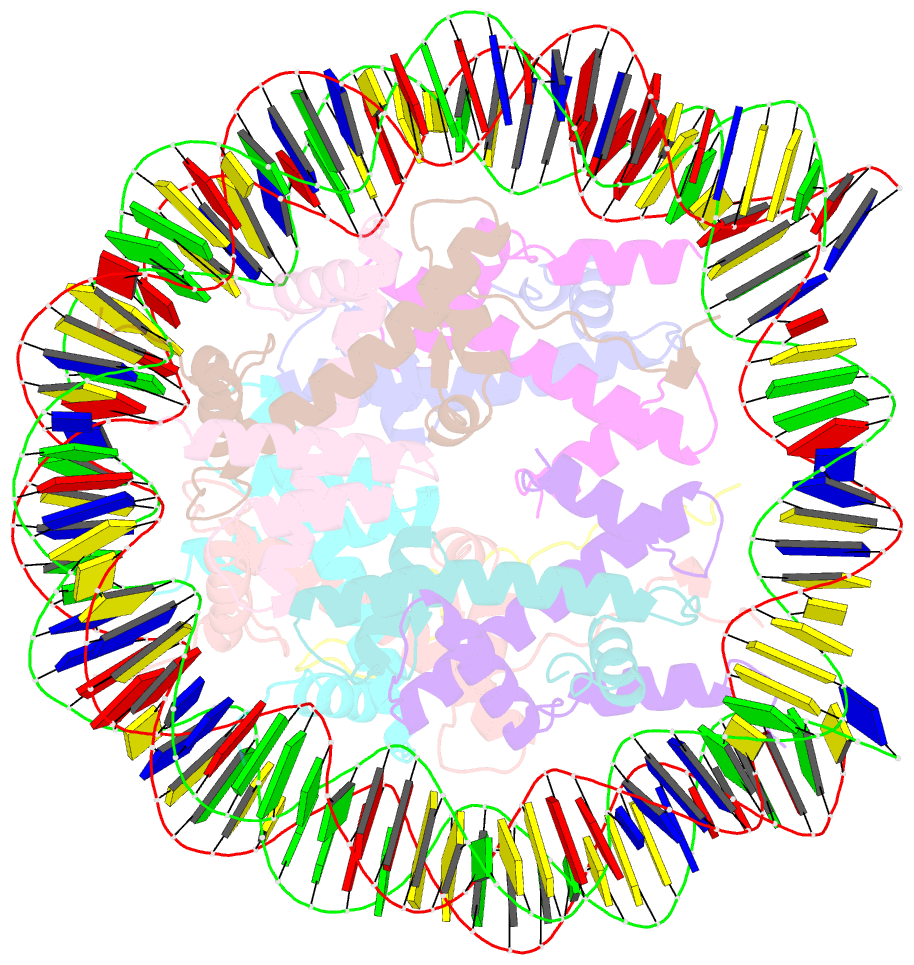
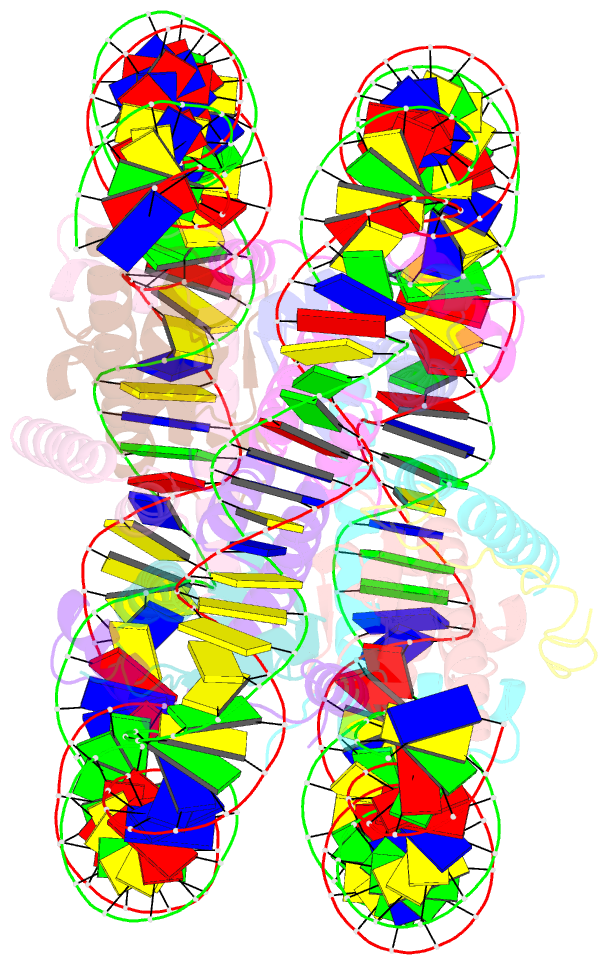
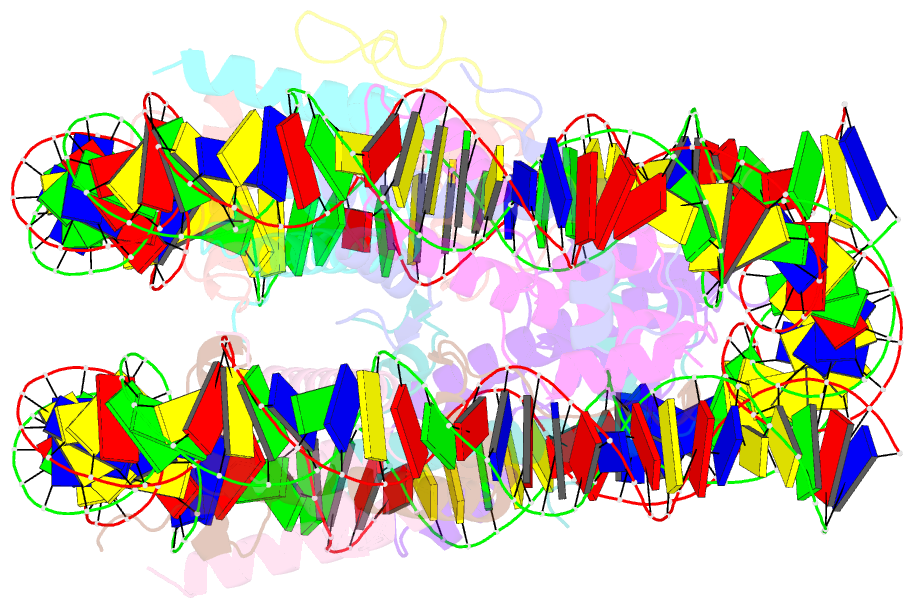
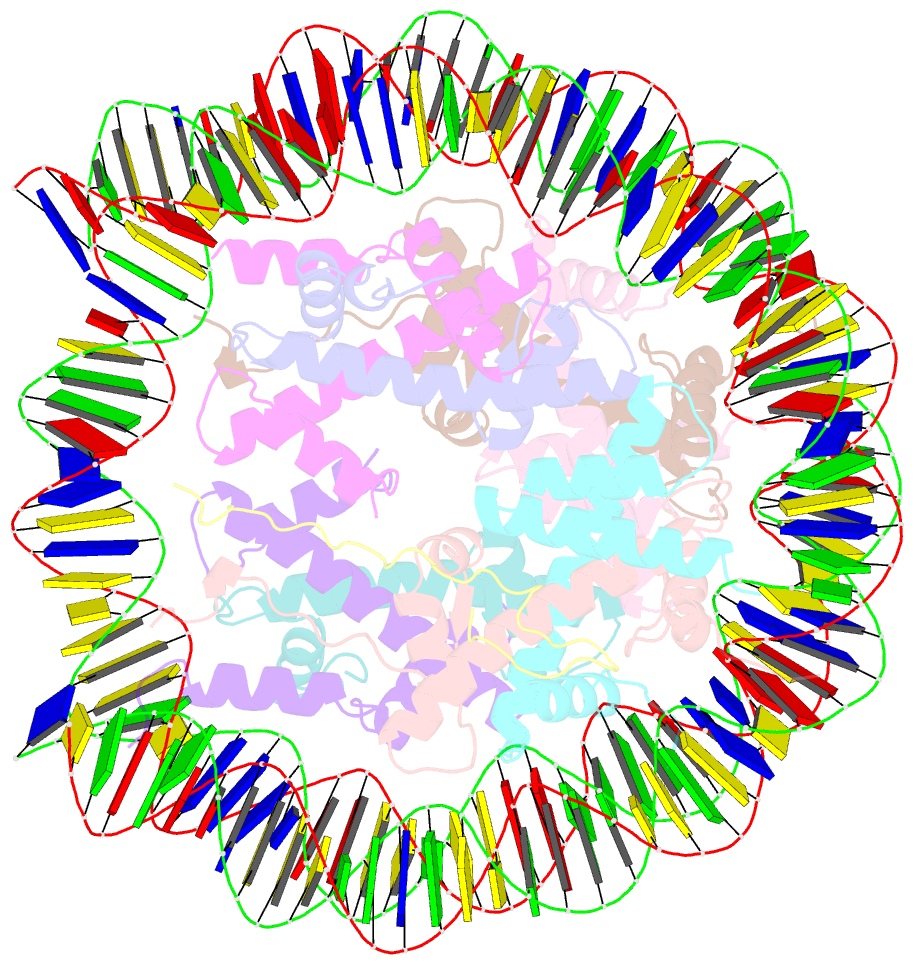
- Summary
- cryo-EM structure of dnmt3a1 udr in complex with h2ak119ub-nucleosome
- Reference
- Gretarsson KH, Abini-Agbomson S, Gloor SL, Weinberg DN, McCuiston JL, Kumary VUS, Hickman AR, Sahu V, Lee R, Xu X, Lipieta N, Flashner S, Adeleke OA, Popova IK, Taylor HF, Noll K, Windham CL, Maryanski DN, Venters BJ, Nakagawa H, Keogh MC, Armache KJ, Lu C (2024): "Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch." Sci Adv, 10, eadp0975. doi: 10.1126/sciadv.adp0975.
- Abstract
- During tumor development, promoter CpG islands that are normally silenced by Polycomb repressive complexes (PRCs) become DNA-hypermethylated. The molecular mechanism by which de novo DNA methyltransferase(s) [DNMT(s)] catalyze CpG methylation at PRC-regulated regions remains unclear. Here, we report a cryo-electron microscopy structure of the DNMT3A long isoform (DNMT3A1) amino-terminal region in complex with a nucleosome carrying PRC1-mediated histone H2A lysine-119 monoubiquitination (H2AK119Ub). We identify regions within the DNMT3A1 amino terminus that bind H2AK119Ub and the nucleosome acidic patch. This bidentate interaction is required for effective DNMT3A1 engagement with H2AK119Ub-modified chromatin in cells. Further, aberrant redistribution of DNMT3A1 to Polycomb target genes recapitulates the cancer-associated DNA hypermethylation signature and inhibits their transcriptional activation during cell differentiation. This effect is rescued by disruption of the DNMT3A1-acidic patch interaction. Together, our analyses reveal a binding interface critical for mediating promoter CpG island DNA hypermethylation, a major molecular hallmark of cancer.