Summary information and primary citation
- PDB-id
- 8v6h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication, transferase-DNA-RNA
- Method
- cryo-EM (11.11 Å)
- Summary
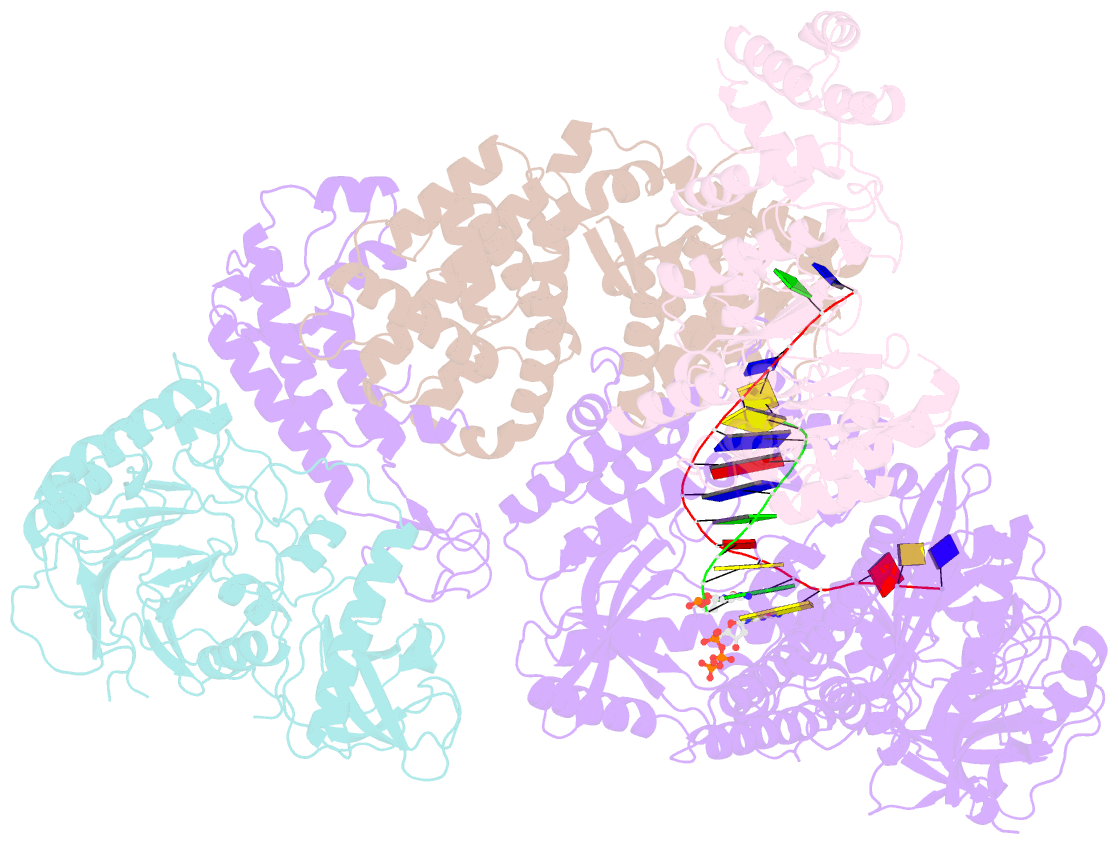
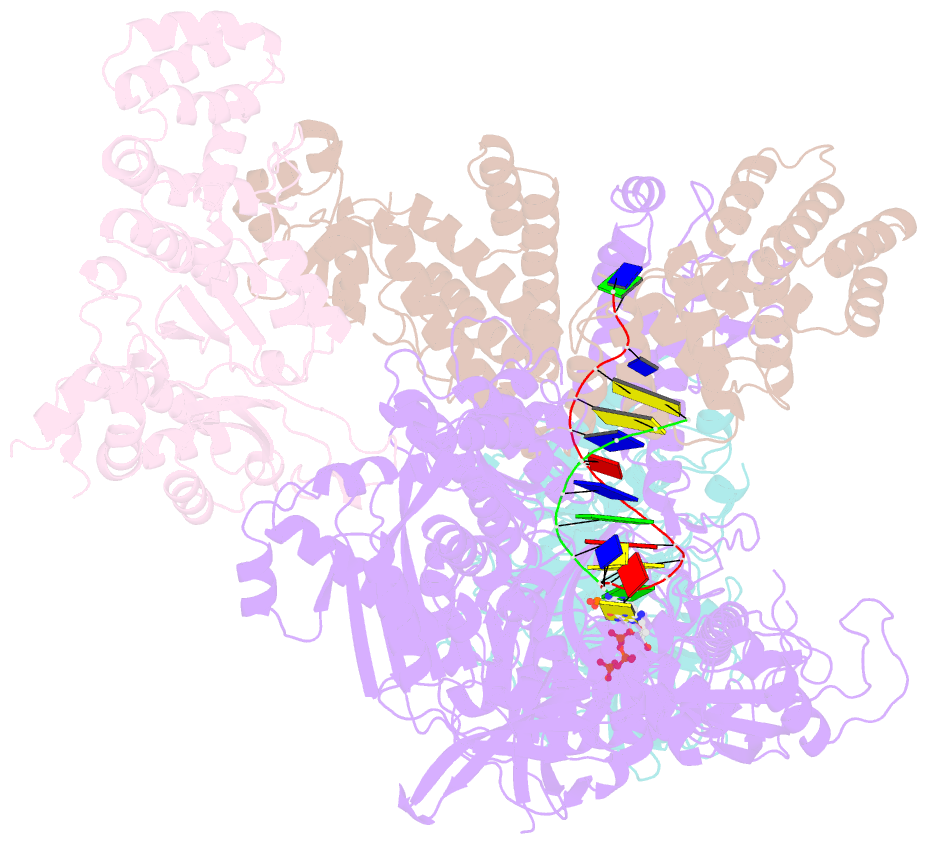
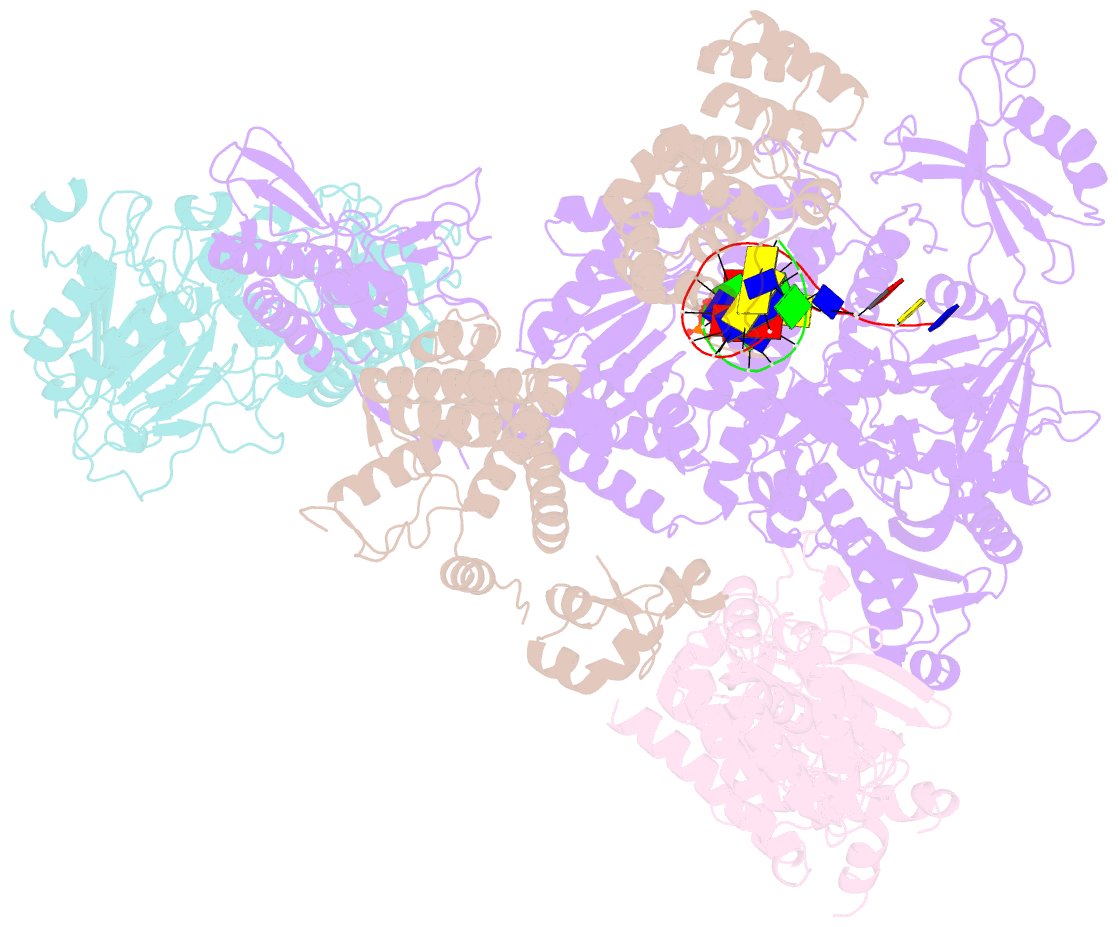
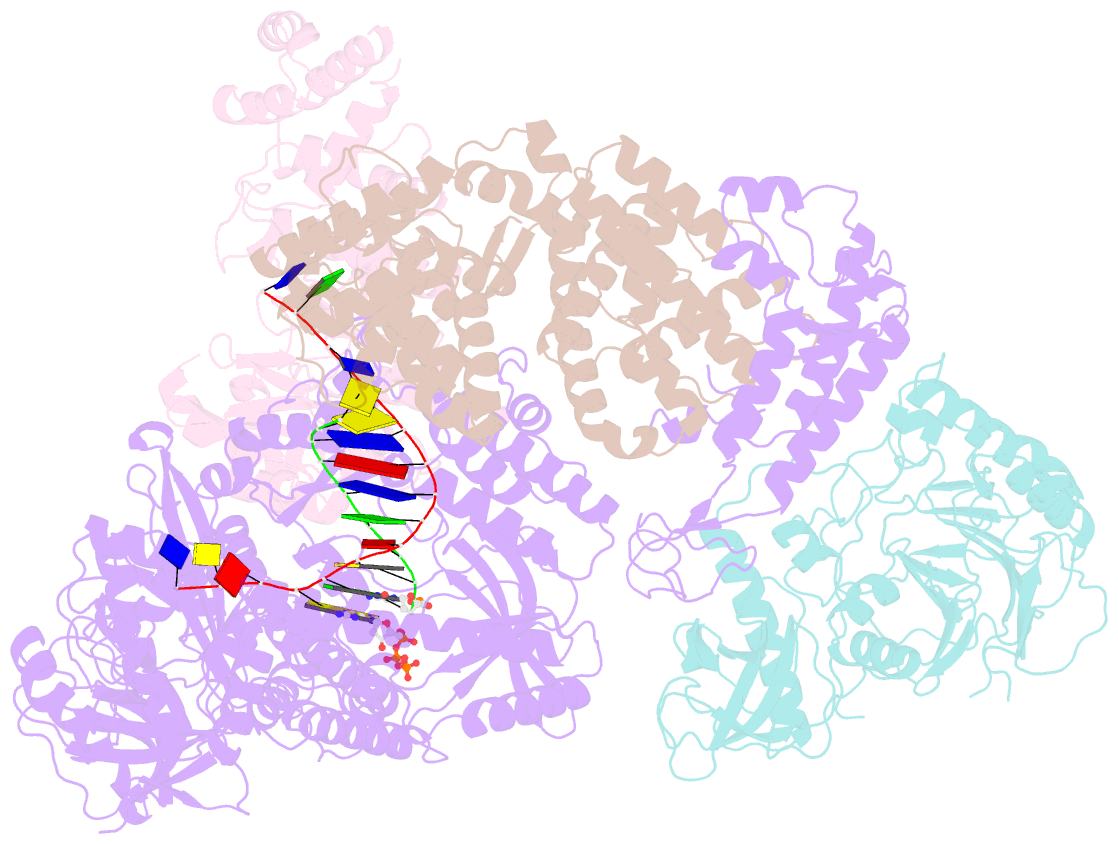
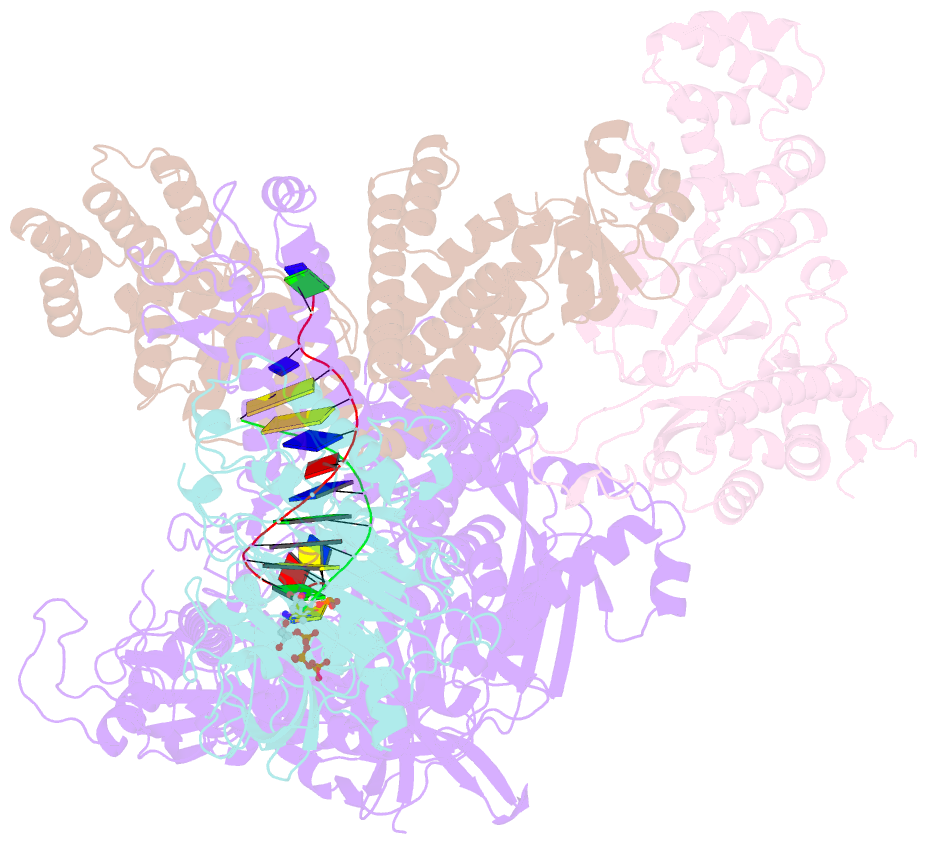
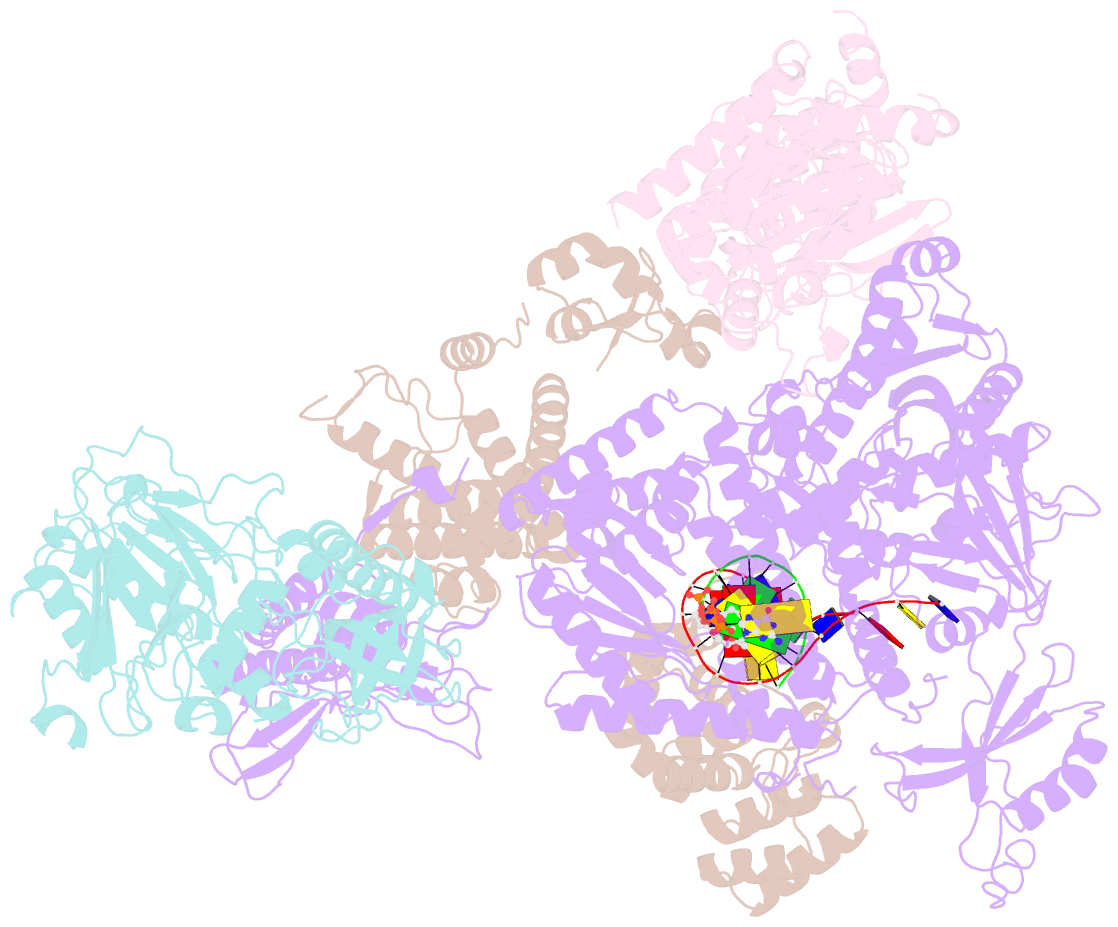
- DNA initiation complex (configuration 2) of xenopus laevis DNA polymerase alpha-primase
- Reference
- Mullins EA, Salay LE, Durie CL, Bradley NP, Jackman JE, Ohi MD, Chazin WJ, Eichman BF (2024): "A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase." Nat.Struct.Mol.Biol., 31, 777-790. doi: 10.1038/s41594-024-01227-4.
- Abstract
- The mechanism by which polymerase α-primase (polα-primase) synthesizes chimeric RNA-DNA primers of defined length and composition, necessary for replication fidelity and genome stability, is unknown. Here, we report cryo-EM structures of Xenopus laevis polα-primase in complex with primed templates representing various stages of DNA synthesis. Our data show how interaction of the primase regulatory subunit with the primer 5' end facilitates handoff of the primer to polα and increases polα processivity, thereby regulating both RNA and DNA composition. The structures detail how flexibility within the heterotetramer enables synthesis across two active sites and provide evidence that termination of DNA synthesis is facilitated by reduction of polα and primase affinities for the varied conformations along the chimeric primer-template duplex. Together, these findings elucidate a critical catalytic step in replication initiation and provide a comprehensive model for primer synthesis by polα-primase.