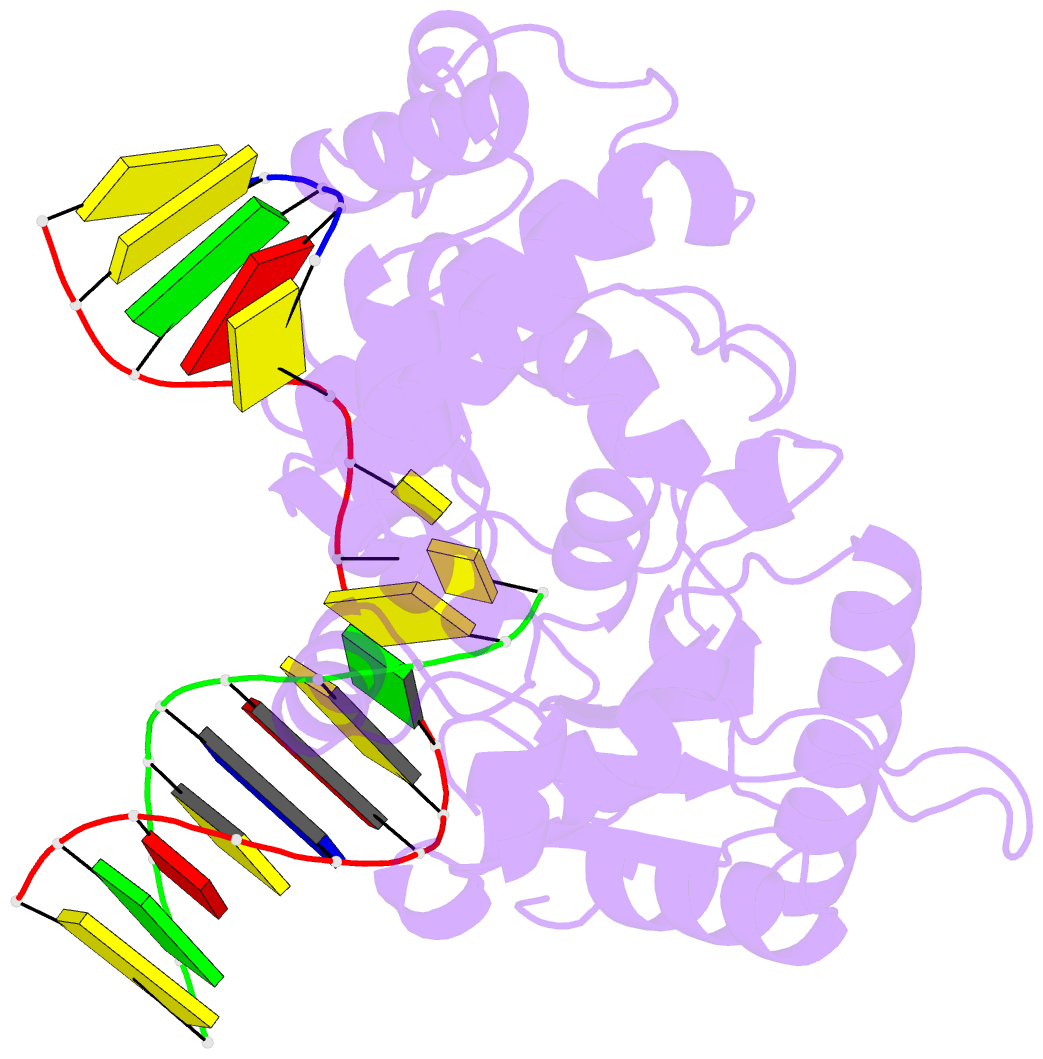
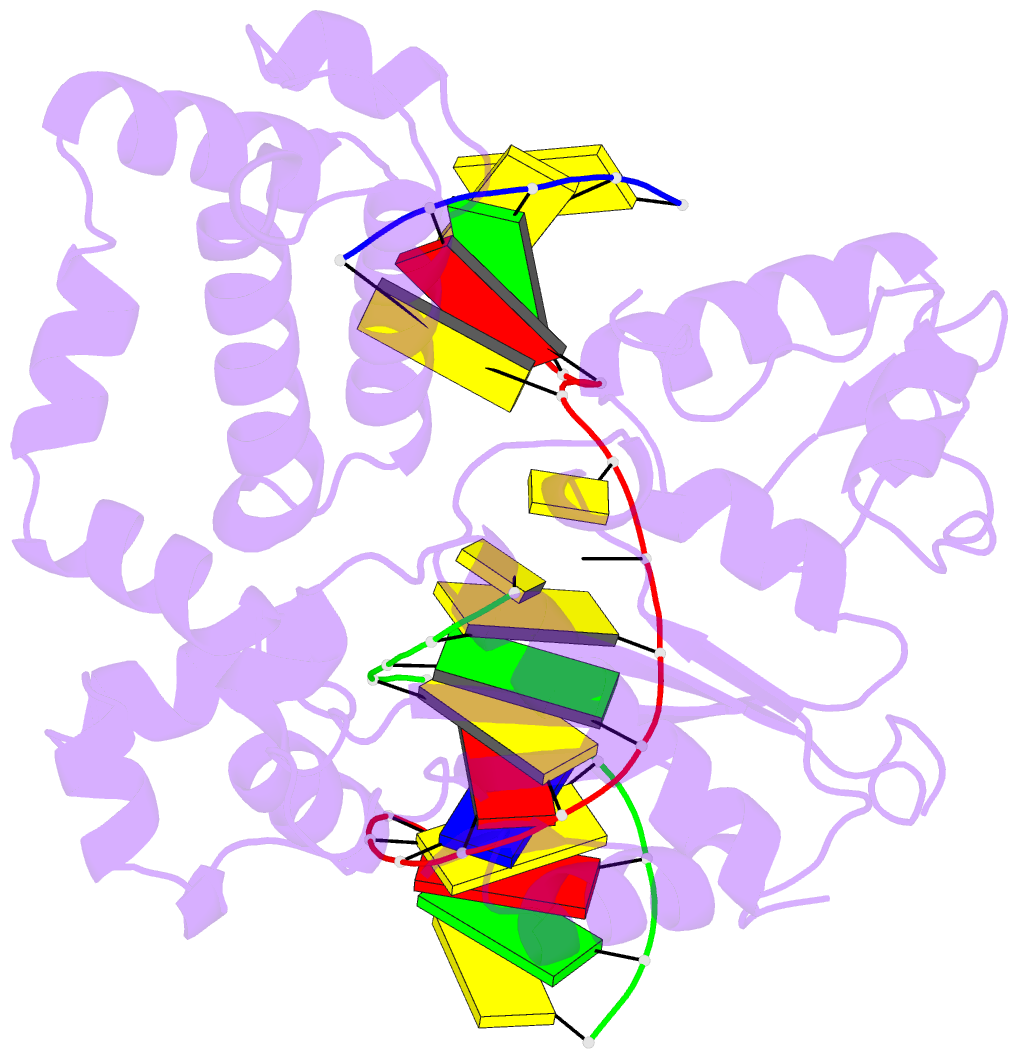
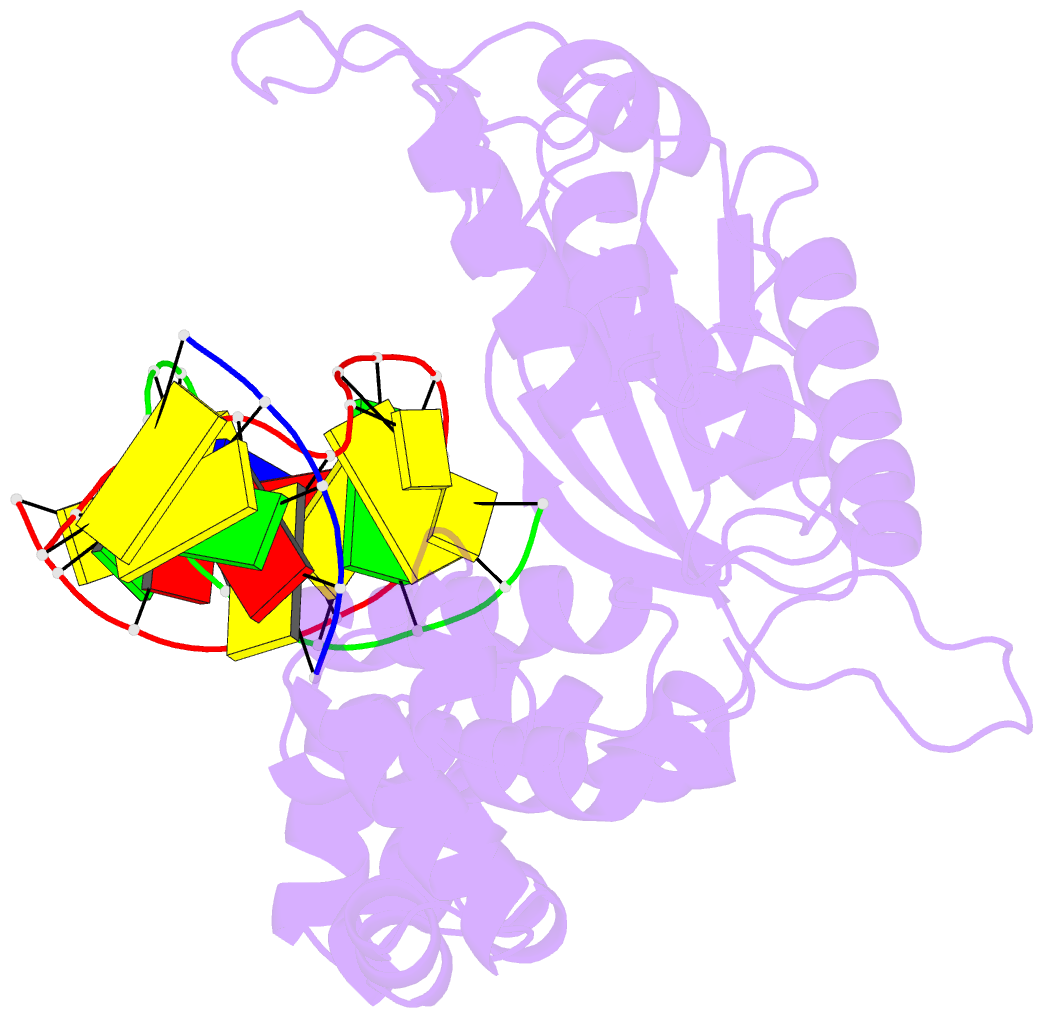
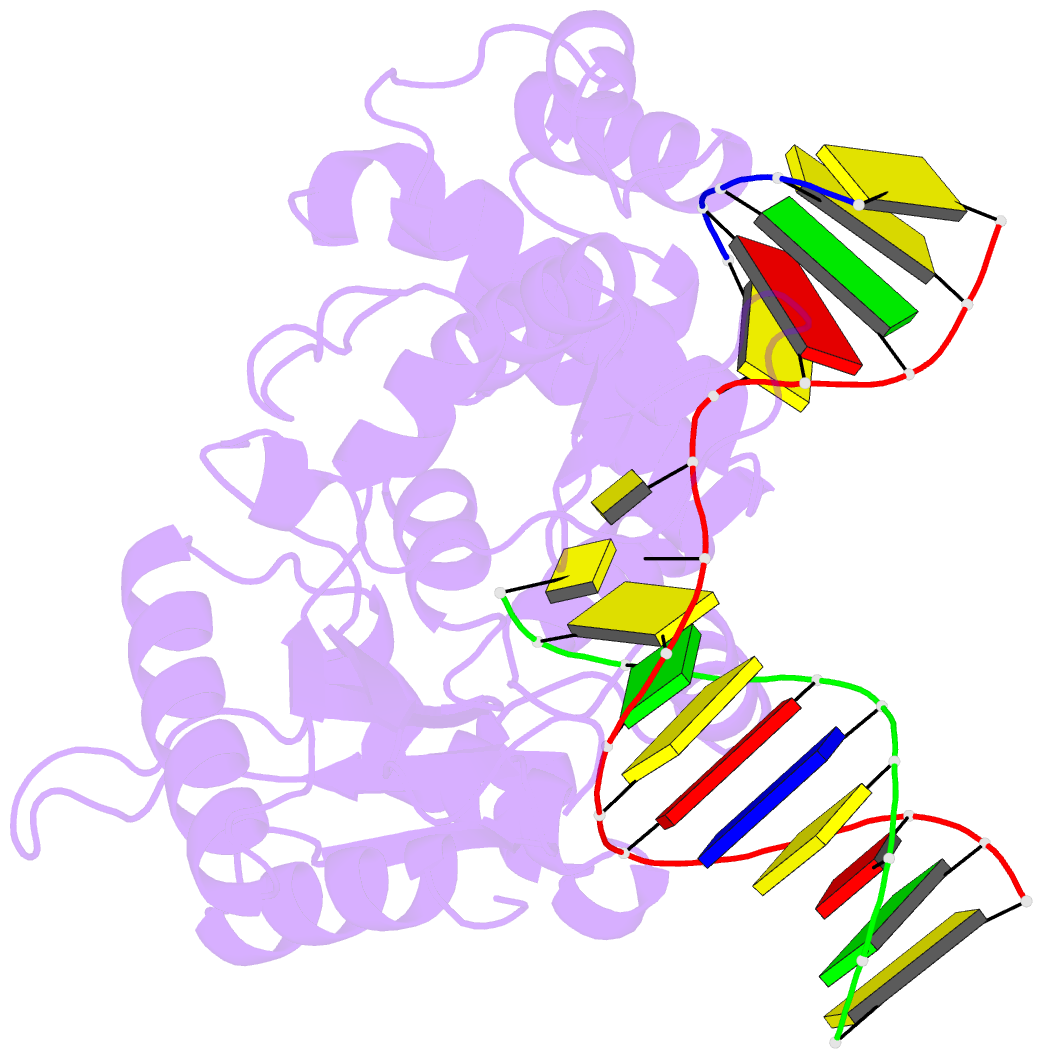
Summary information and primary citation
- PDB-id
- 8vf8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (1.98 Å)
- Summary
- Binary DNA polymerase beta bound to DNA containing primer terminal dc base-paired with fapydg
- Reference
- Gao S, Oden PN, Ryan BJ, Yang H, Freudenthal BD, Greenberg MM (2024): "Biochemical and structural characterization of Fapy•dG replication by Human DNA polymerase beta." Nucleic Acids Res., 52, 5392-5405. doi: 10.1093/nar/gkae277.
- Abstract
- N6-(2-deoxy-α,β-d-erythro-pentofuranosyl)-2,6-diamino-4-hydroxy-5-formamido-pyrimidine (Fapy•dG) is formed from a common intermediate and in comparable amounts to the well-studied mutagenic DNA lesion 8-oxo-7,8-dihydro-2'-deoxyguanosine (8-OxodGuo). Fapy•dG preferentially gives rise to G → T transversions and G → A transitions. However, the molecular basis by which Fapy•dG is processed by DNA polymerases during this mutagenic process remains poorly understood. To address this we investigated how DNA polymerase β (Pol β), a model mammalian polymerase, bypasses a templating Fapy•dG, inserts Fapy•dGTP, and extends from Fapy•dG at the primer terminus. When Fapy•dG is present in the template, Pol β incorporates TMP less efficiently than either dCMP or dAMP. Kinetic analysis revealed that Fapy•dGTP is a poor substrate but is incorporated ∼3-times more efficiently opposite dA than dC. Extension from Fapy•dG at the 3'-terminus of a nascent primer is inefficient due to the primer terminus being poorly positioned for catalysis. Together these data indicate that mutagenic bypass of Fapy•dG is likely to be the source of the mutagenic effects of the lesion and not Fapy•dGTP. These experiments increase our understanding of the promutagenic effects of Fapy•dG.