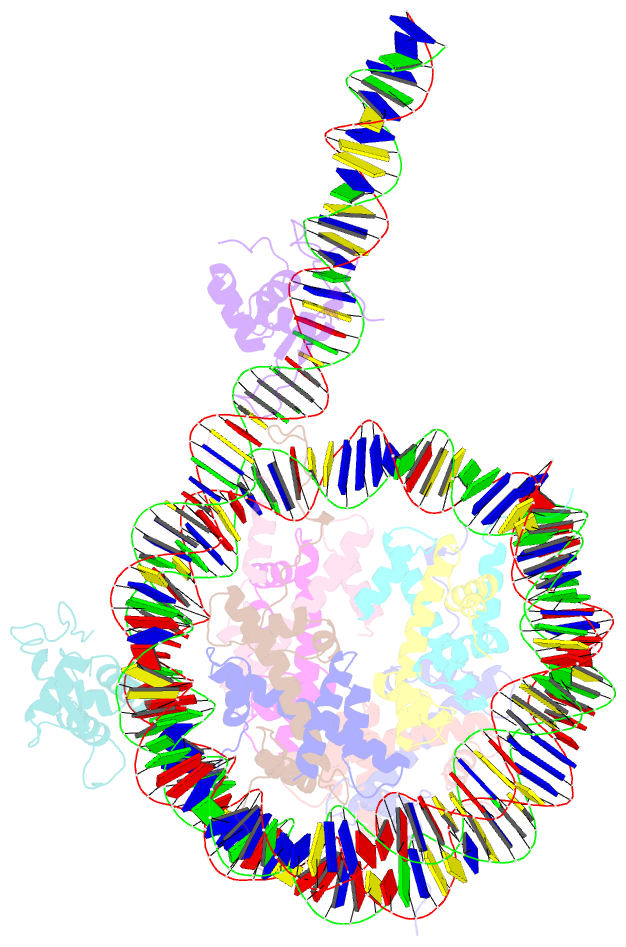
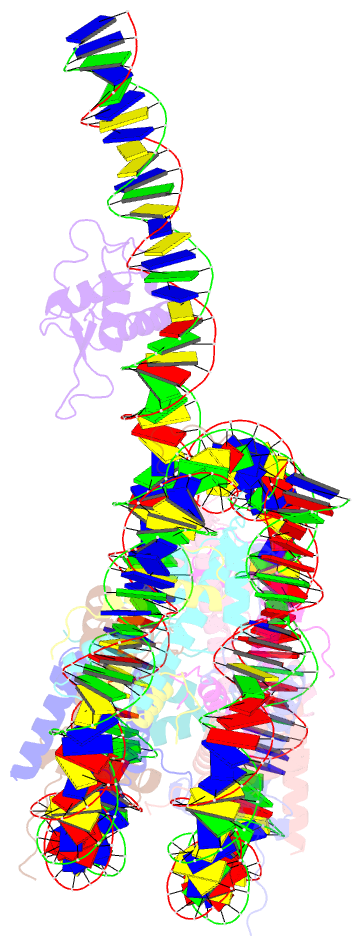
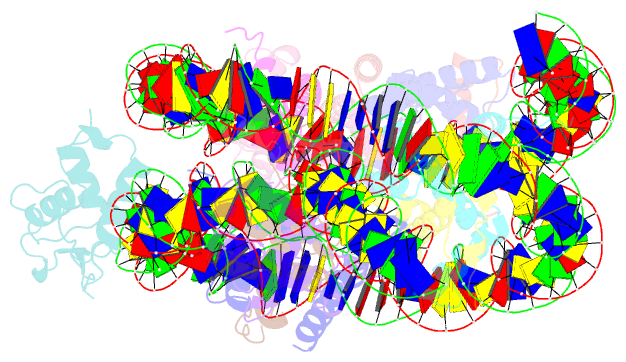
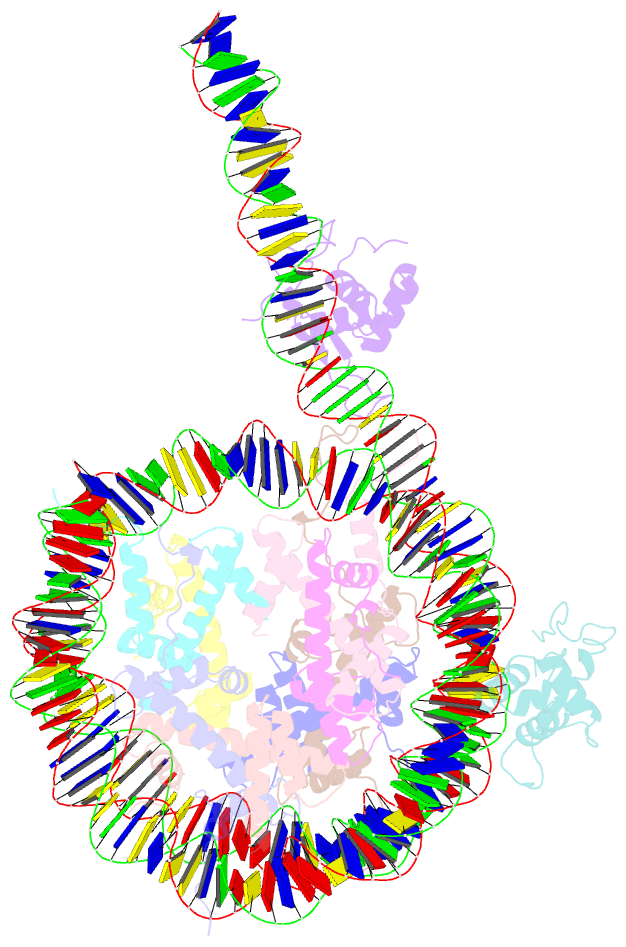
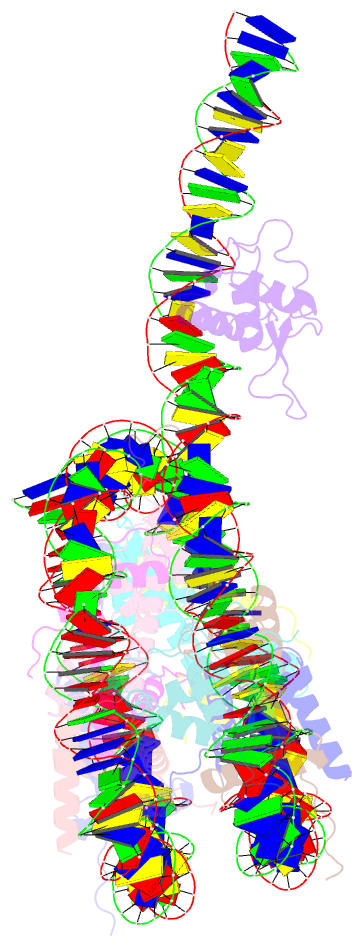
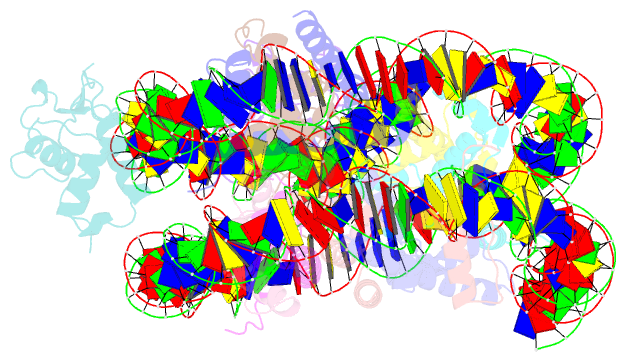
Summary information and primary citation
- PDB-id
- 8vfz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein-DNA
- Method
- cryo-EM (4.1 Å)
- Summary
- cryo-EM structure of foxa1 in complex with albn1 nucleosome (class 2)
- Reference
- Zhou BR, Feng H, Huang F, Zhu I, Portillo-Ledesma S, Shi D, Zaret KS, Schlick T, Landsman D, Wang Q, Bai Y (2024): "Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4." Mol.Cell, 84, 3061. doi: 10.1016/j.molcel.2024.07.016.
- Abstract
- Mouse FOXA1 and GATA4 are prototypes of pioneer factors, initiating liver cell development by binding to the N1 nucleosome in the enhancer of the ALB1 gene. Using cryoelectron microscopy (cryo-EM), we determined the structures of the free N1 nucleosome and its complexes with FOXA1 and GATA4, both individually and in combination. We found that the DNA-binding domains of FOXA1 and GATA4 mainly recognize the linker DNA and an internal site in the nucleosome, respectively, whereas their intrinsically disordered regions interact with the acidic patch on histone H2A-H2B. FOXA1 efficiently enhances GATA4 binding by repositioning the N1 nucleosome. In vivo DNA editing and bioinformatics analyses suggest that the co-binding mode of FOXA1 and GATA4 plays important roles in regulating genes involved in liver cell functions. Our results reveal the mechanism whereby FOXA1 and GATA4 cooperatively bind to the nucleosome through nucleosome repositioning, opening chromatin by bending linker DNA and obstructing nucleosome packing.