Summary information and primary citation
- PDB-id
- 8vm9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-immune system
- Method
- X-ray (2.2 Å)
- Summary
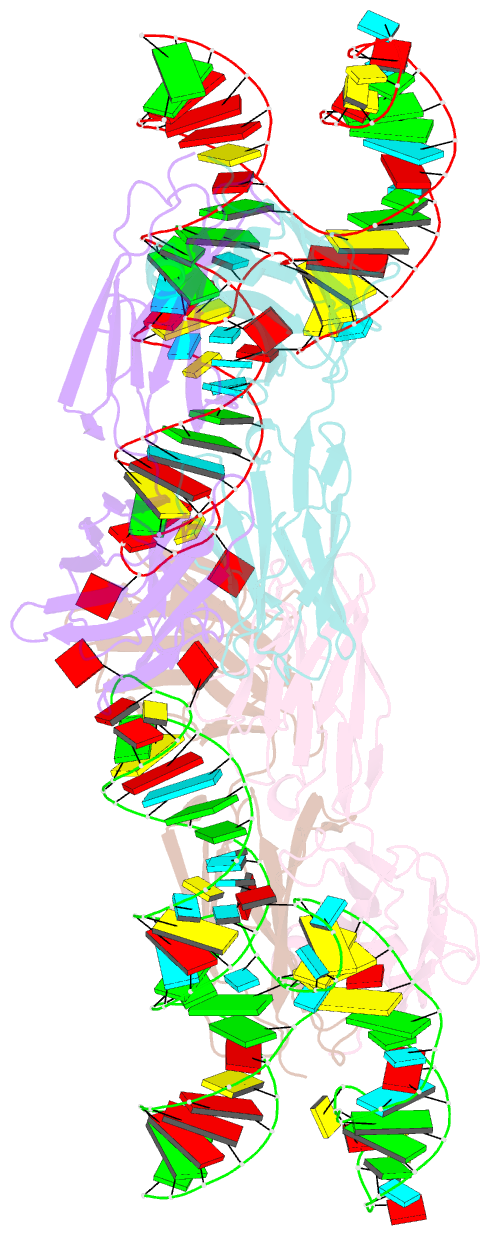
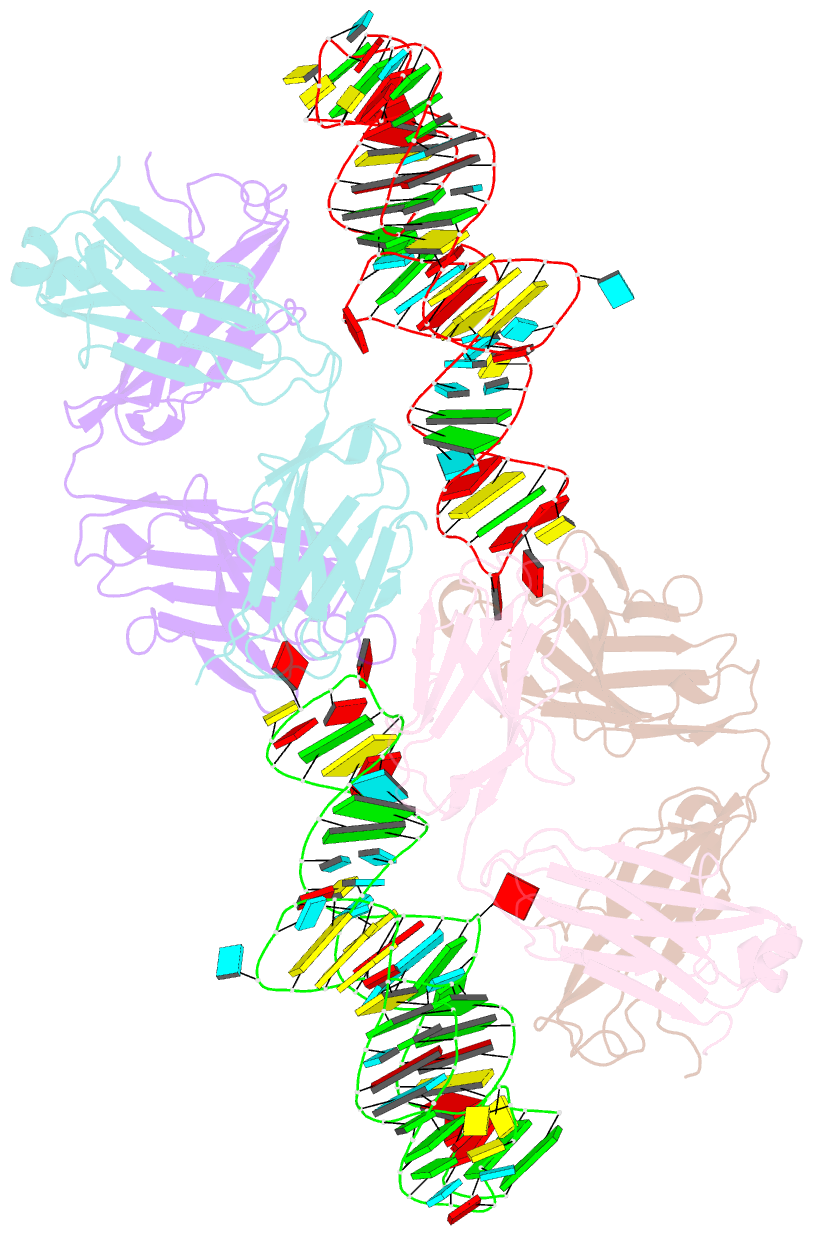
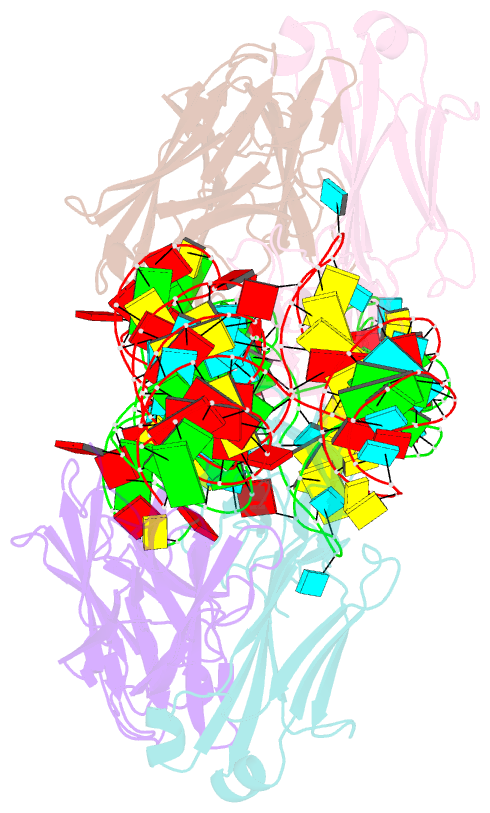
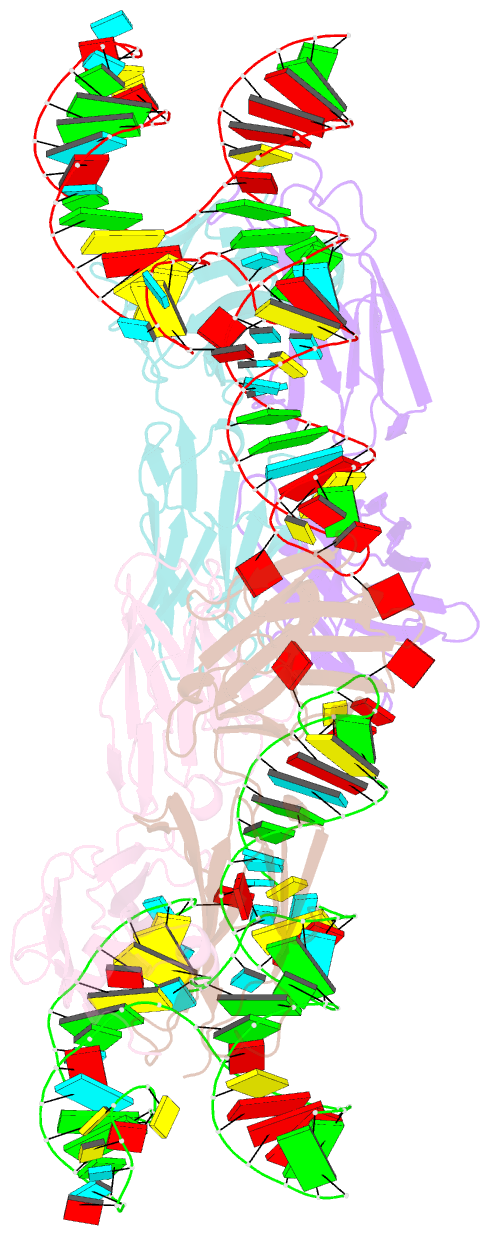
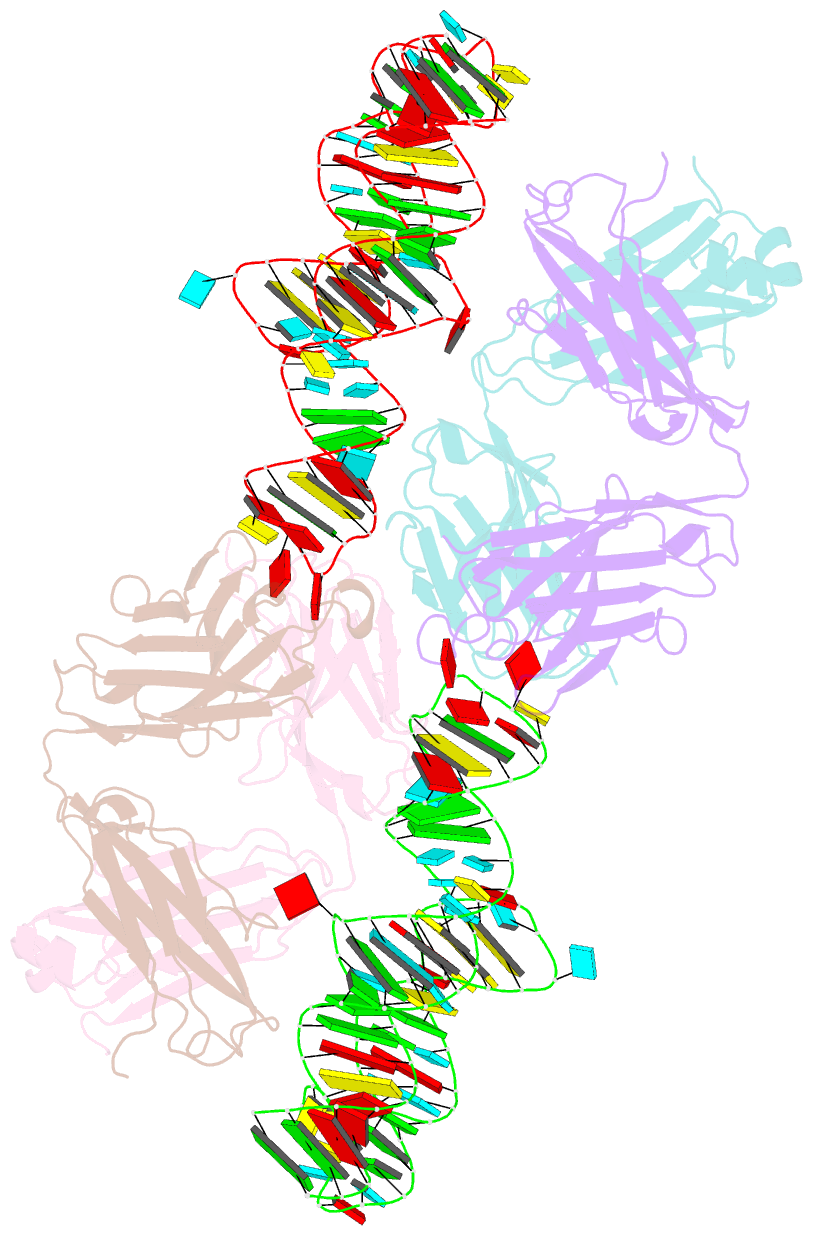
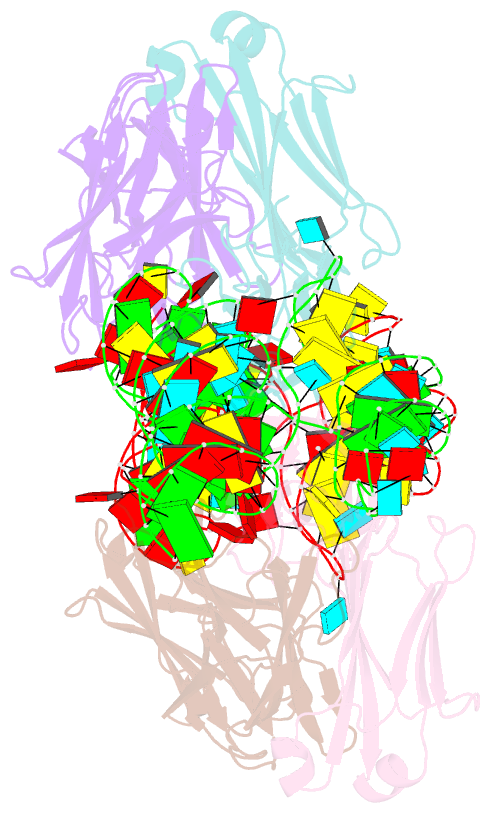
- The crystal structure of rhinovirus b14 RNA replication element sd-loop mutant in complex with fab bl3-6
- Reference
- Das NK, Vogt J, Patel A, Banna HA, Koirala D (2024): "Structural basis for a highly conserved RNA-mediated enteroviral genome replication." Nucleic Acids Res., 52, 11218-11233. doi: 10.1093/nar/gkae627.
- Abstract
- Enteroviruses contain conserved RNA structures at the extreme 5' end of their genomes that recruit essential proteins 3CD and PCBP2 to promote genome replication. However, the high-resolution structures and mechanisms of these replication-linked RNAs (REPLRs) are limited. Here, we determined the crystal structures of the coxsackievirus B3 and rhinoviruses B14 and C15 REPLRs at 1.54, 2.2 and 2.54 Å resolution, revealing a highly conserved H-type four-way junction fold with co-axially stacked sA-sD and sB-sC helices that are stabilized by a long-range A•C•U base-triple. Such conserved features observed in the crystal structures also allowed us to predict the models of several other enteroviral REPLRs using homology modeling, which generated models almost identical to the experimentally determined structures. Moreover, our structure-guided binding studies with recombinantly purified full-length human PCBP2 showed that two previously proposed binding sites, the sB-loop and 3' spacer, reside proximally and bind a single PCBP2. Additionally, the DNA oligos complementary to the 3' spacer, the high-affinity PCBP2 binding site, abrogated its interactions with enteroviral REPLRs, suggesting the critical roles of this single-stranded region in recruiting PCBP2 for enteroviral genome replication and illuminating the promising prospects of developing therapeutics against enteroviral infections targeting this replication platform.