Summary information and primary citation
- PDB-id
- 8w7m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication
- Method
- cryo-EM (4.12 Å)
- Summary
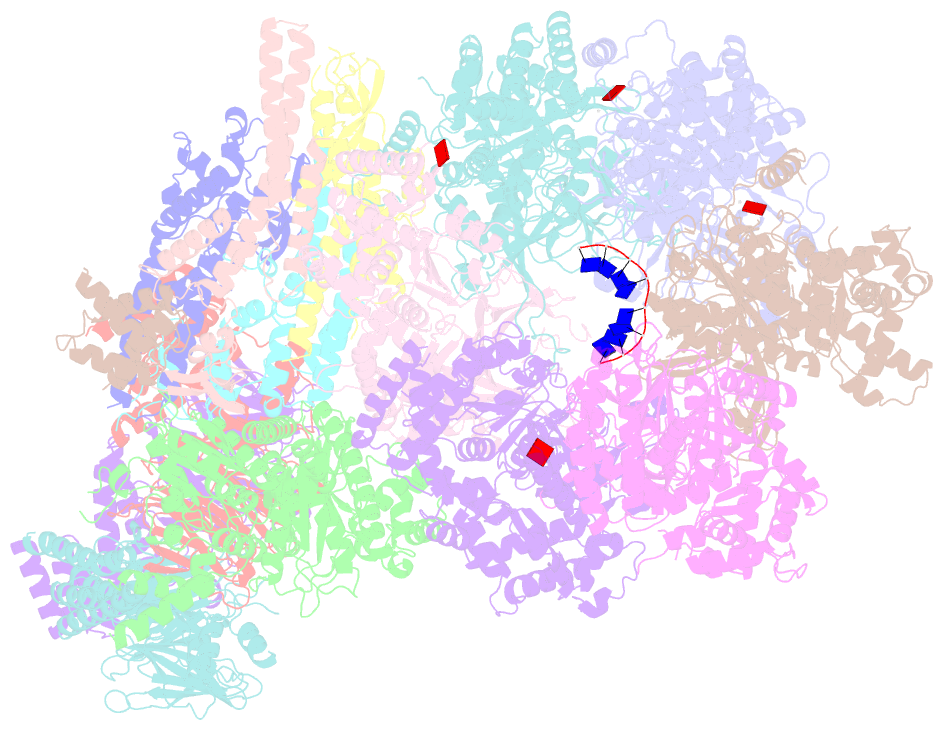
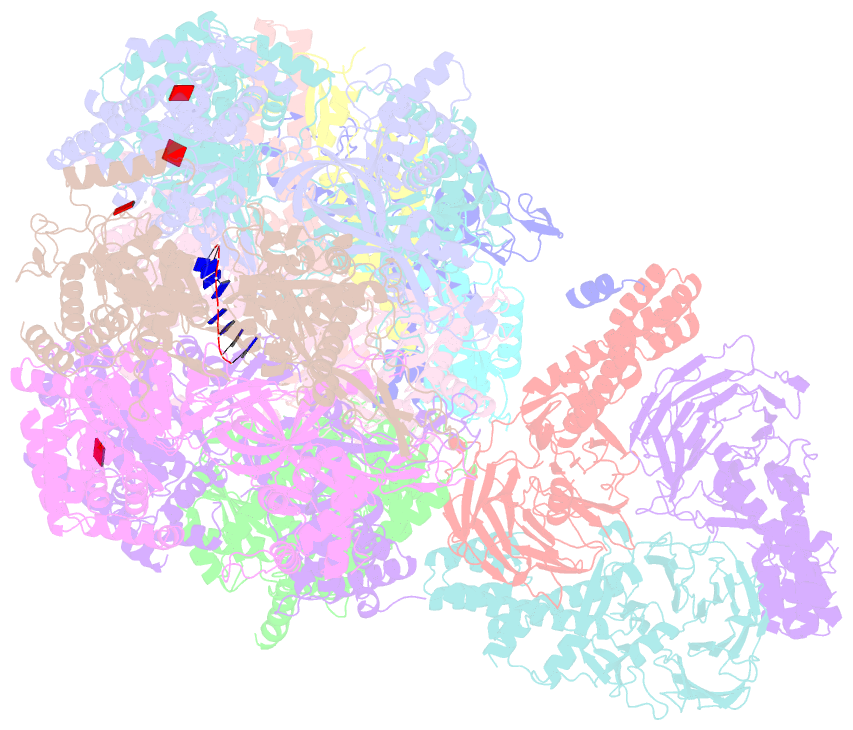
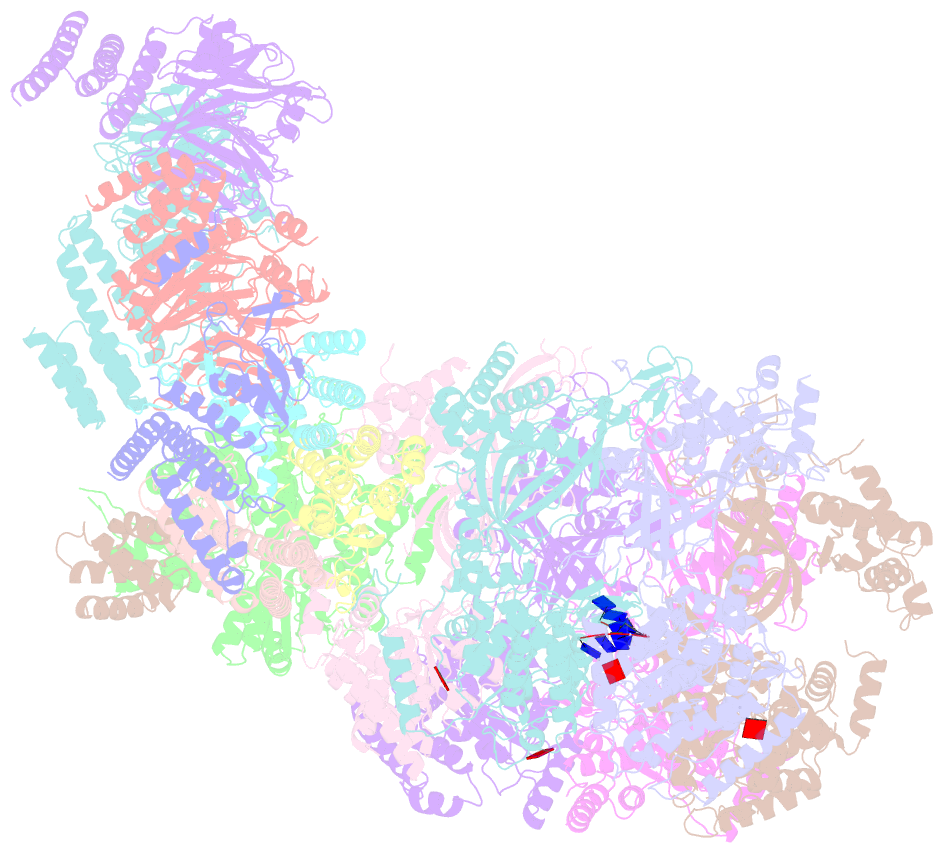
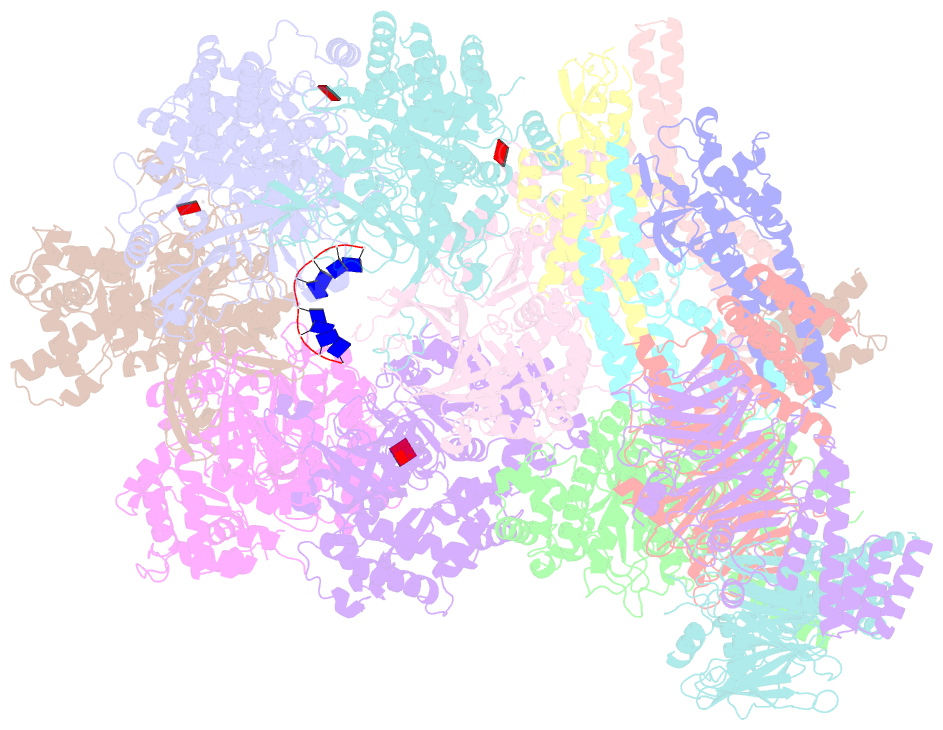
- Yeast replisome in state v
- Reference
- Xu Z, Feng J, Yu D, Huo Y, Ma X, Lam WH, Liu Z, Li XD, Ishibashi T, Dang S, Zhai Y (2023): "Synergism between CMG helicase and leading strand DNA polymerase at replication fork." Nat Commun, 14, 5849. doi: 10.1038/s41467-023-41506-0.
- Abstract
- The replisome that replicates the eukaryotic genome consists of at least three engines: the Cdc45-MCM-GINS (CMG) helicase that separates duplex DNA at the replication fork and two DNA polymerases, one on each strand, that replicate the unwound DNA. Here, we determined a series of cryo-electron microscopy structures of a yeast replisome comprising CMG, leading-strand polymerase Polε and three accessory factors on a forked DNA. In these structures, Polε engages or disengages with the motor domains of the CMG by occupying two alternative positions, which closely correlate with the rotational movement of the single-stranded DNA around the MCM pore. During this process, the polymerase remains stably coupled to the helicase using Psf1 as a hinge. This synergism is modulated by a concerted rearrangement of ATPase sites to drive DNA translocation. The Polε-MCM coupling is not only required for CMG formation to initiate DNA replication but also facilitates the leading-strand DNA synthesis mediated by Polε. Our study elucidates a mechanism intrinsic to the replisome that coordinates the activities of CMG and Polε to negotiate any roadblocks, DNA damage, and epigenetic marks encountered during translocation along replication forks.