Summary information and primary citation
- PDB-id
- 8w9f; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (4.4 Å)
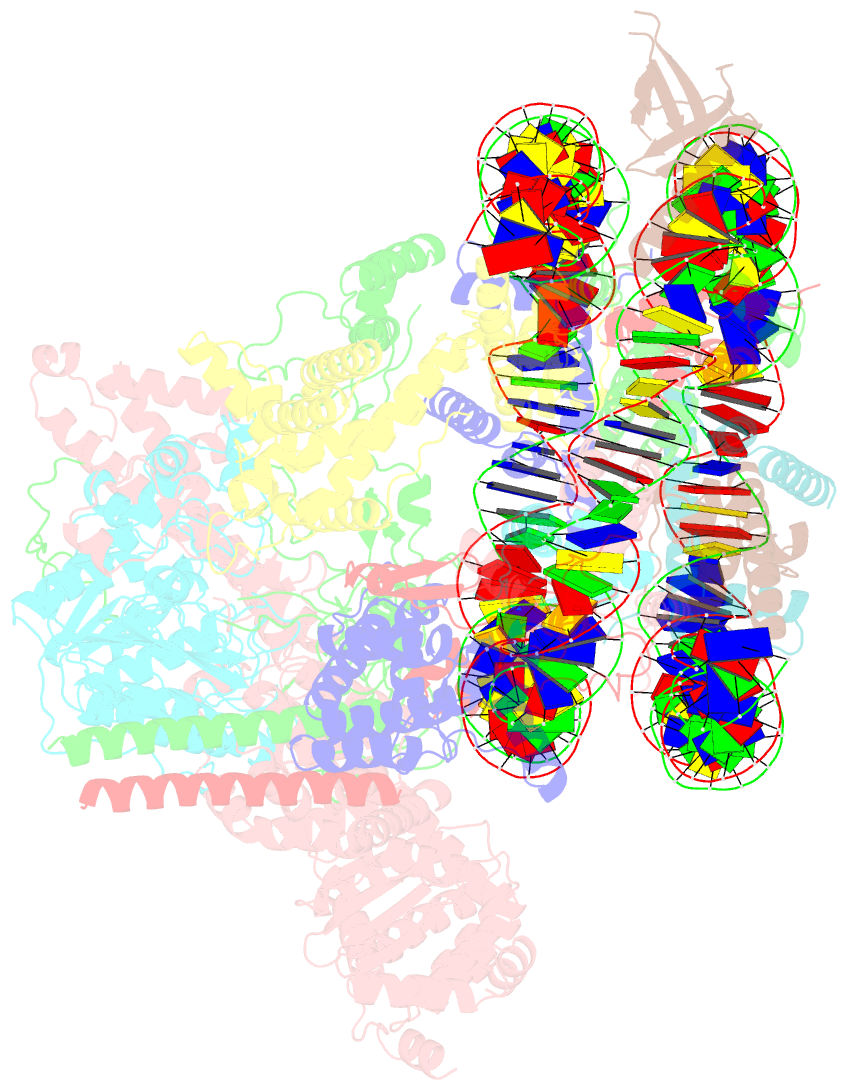
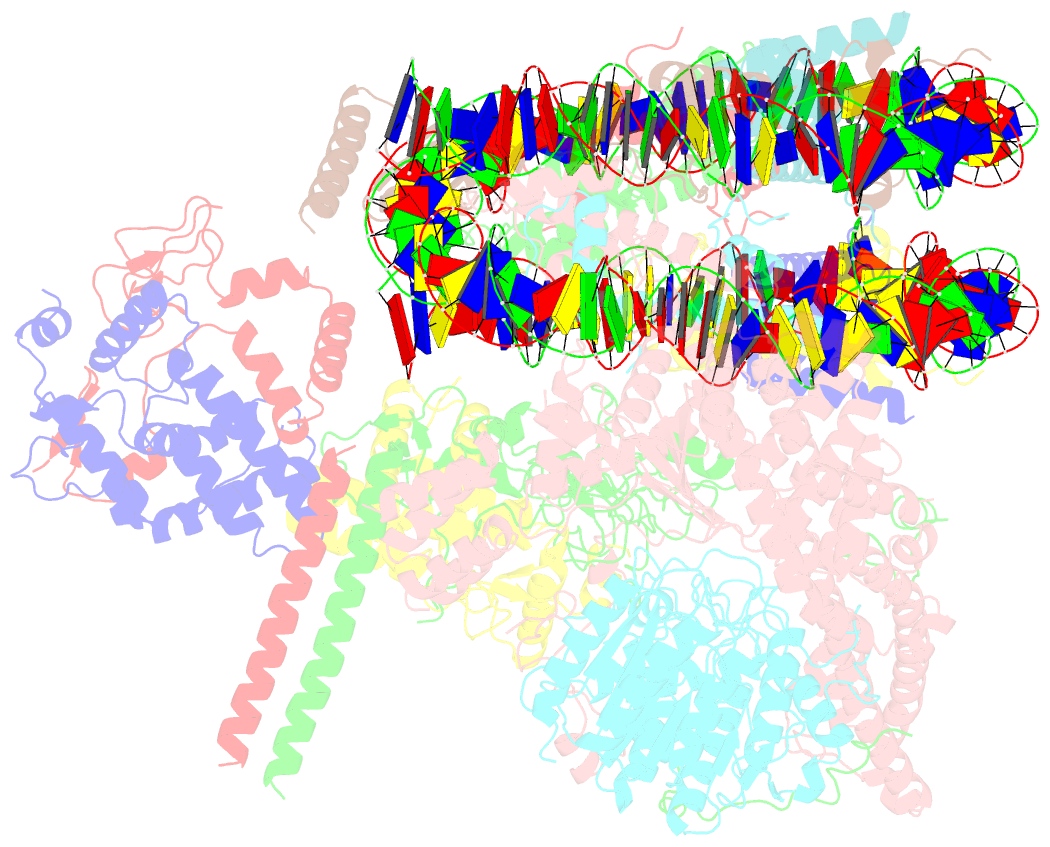
- Summary
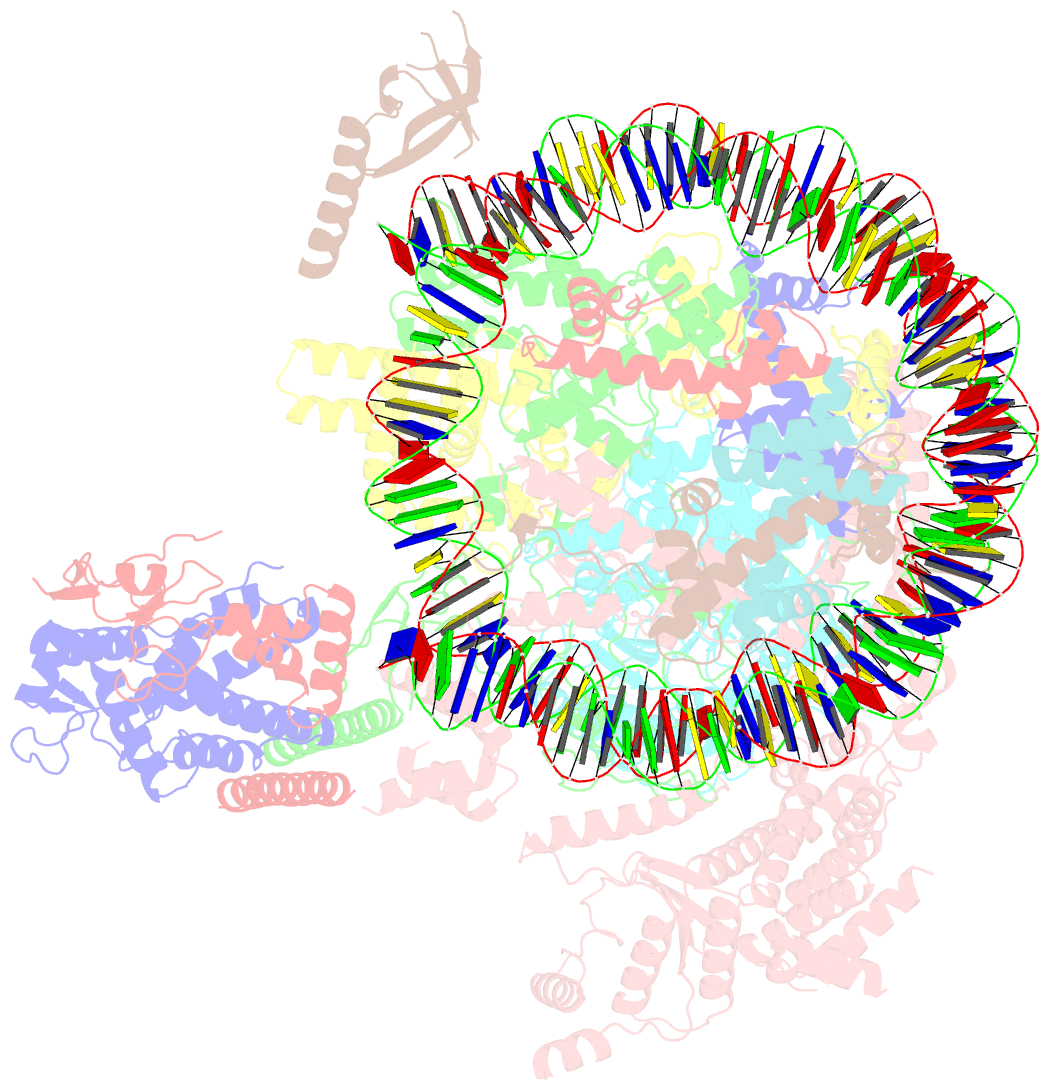
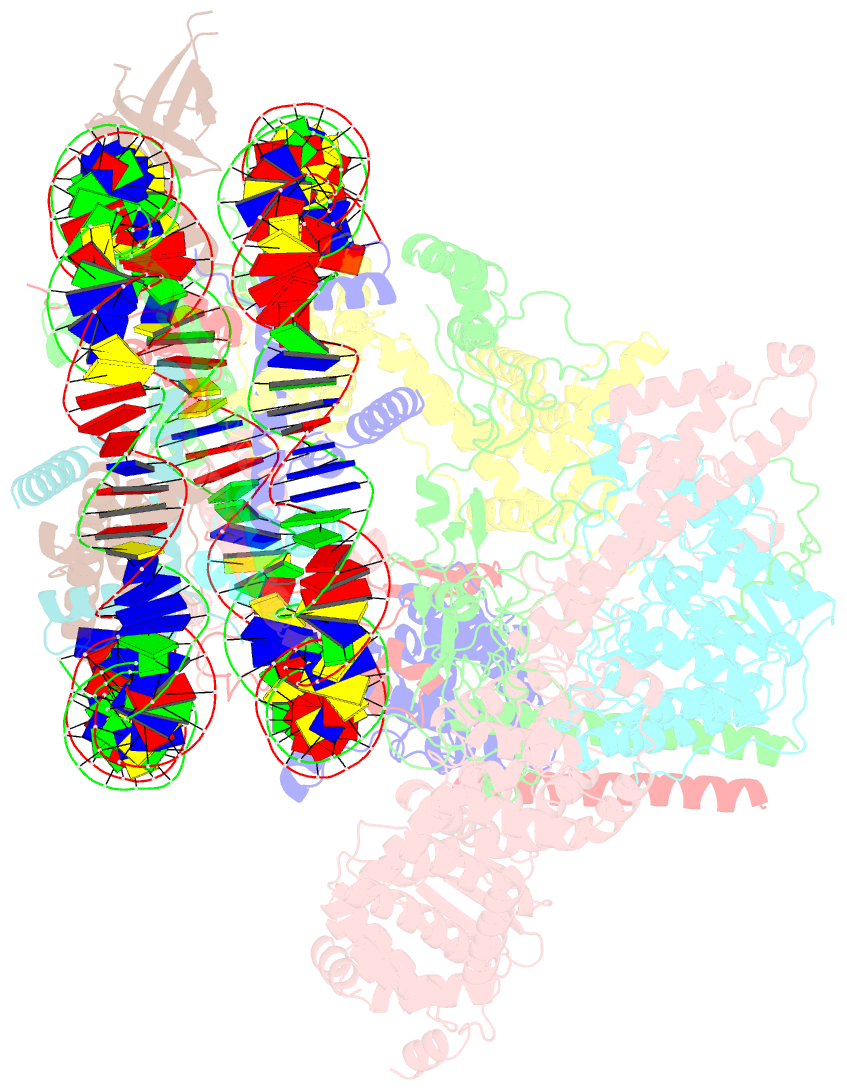
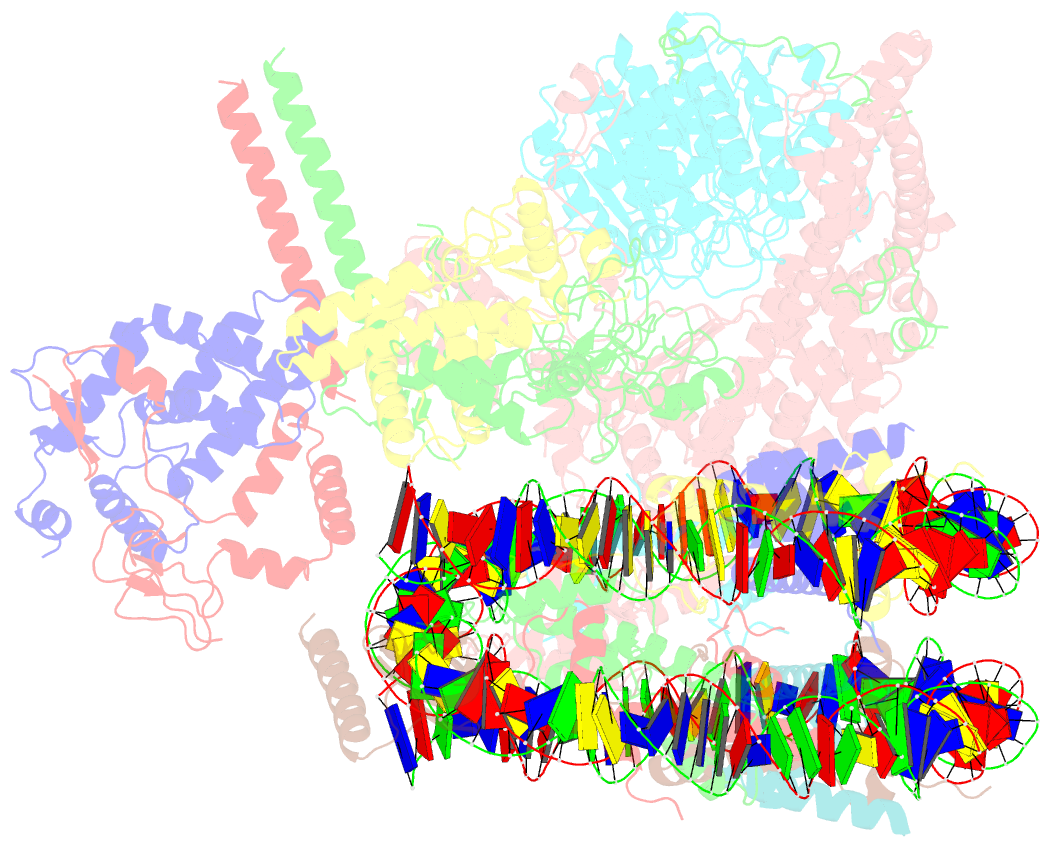
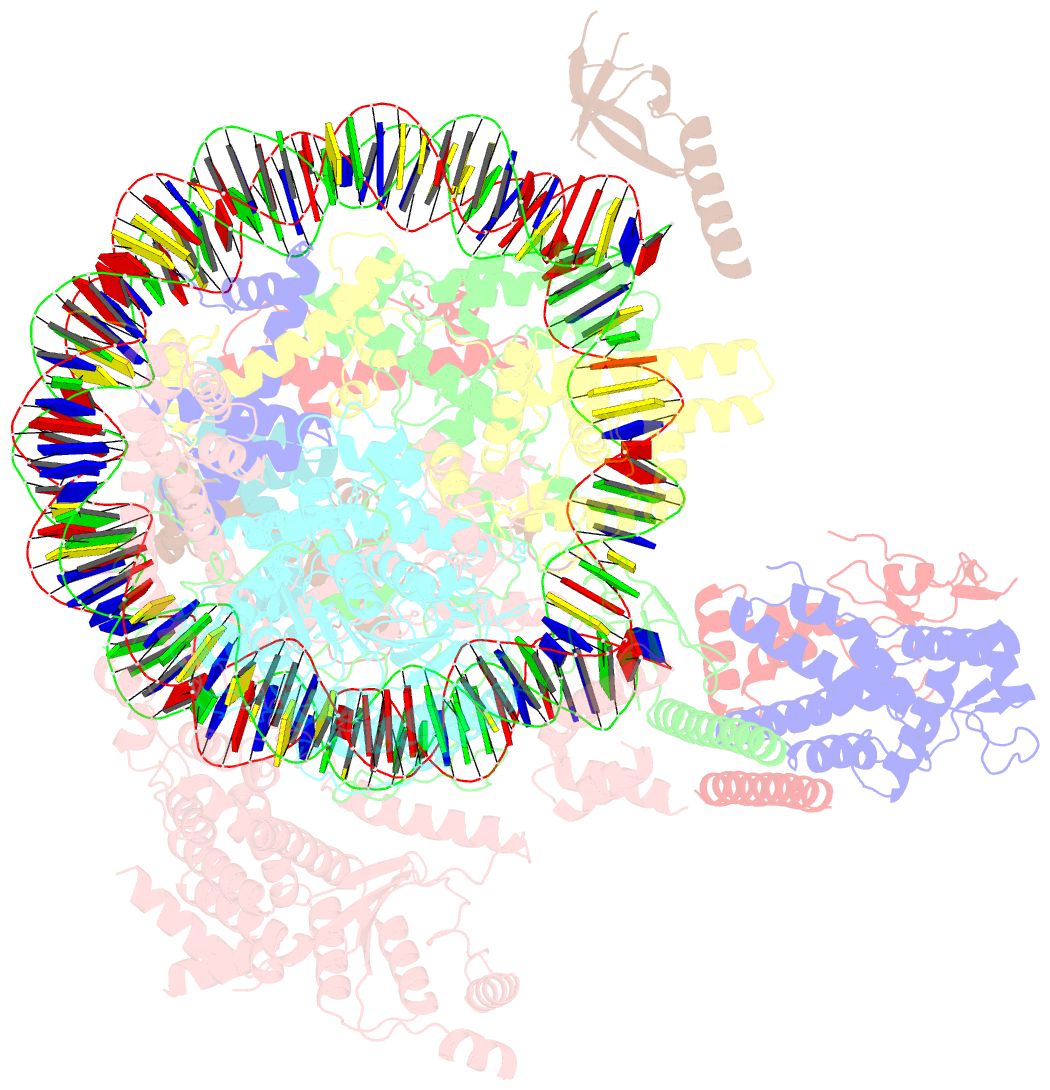
- cryo-EM structure of the rpd3s-nucleosome complex from budding yeast in state 3
- Reference
- Wang C, Chu C, Guo Z, Zhan X (2024): "Structures and dynamics of Rpd3S complex bound to nucleosome." Sci Adv, 10, eadk7678. doi: 10.1126/sciadv.adk7678.
- Abstract
- The Rpd3S complex plays a pivotal role in facilitating local histone deacetylation in the transcribed regions to suppress intragenic transcription initiation. Here, we present the cryo-electron microscopy structures of the budding yeast Rpd3S complex in both its apo and three nucleosome-bound states at atomic resolutions, revealing the exquisite architecture of Rpd3S to well accommodate a mononucleosome without linker DNA. The Rpd3S core, containing a Sin3 Lobe and two NB modules, is a rigid complex and provides three positive-charged anchors (Sin3_HCR and two Rco1_NIDs) to connect nucleosomal DNA. In three nucleosome-bound states, the Rpd3S core exhibits three distinct orientations relative to the nucleosome, assisting the sector-shaped deacetylase Rpd3 to locate above the SHL5-6, SHL0-1, or SHL2-3, respectively. Our work provides a structural framework that reveals a dynamic working model for the Rpd3S complex to engage diverse deacetylation sites.