Summary information and primary citation
- PDB-id
- 8wg5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- nuclear protein
- Method
- cryo-EM (3.05 Å)
- Summary
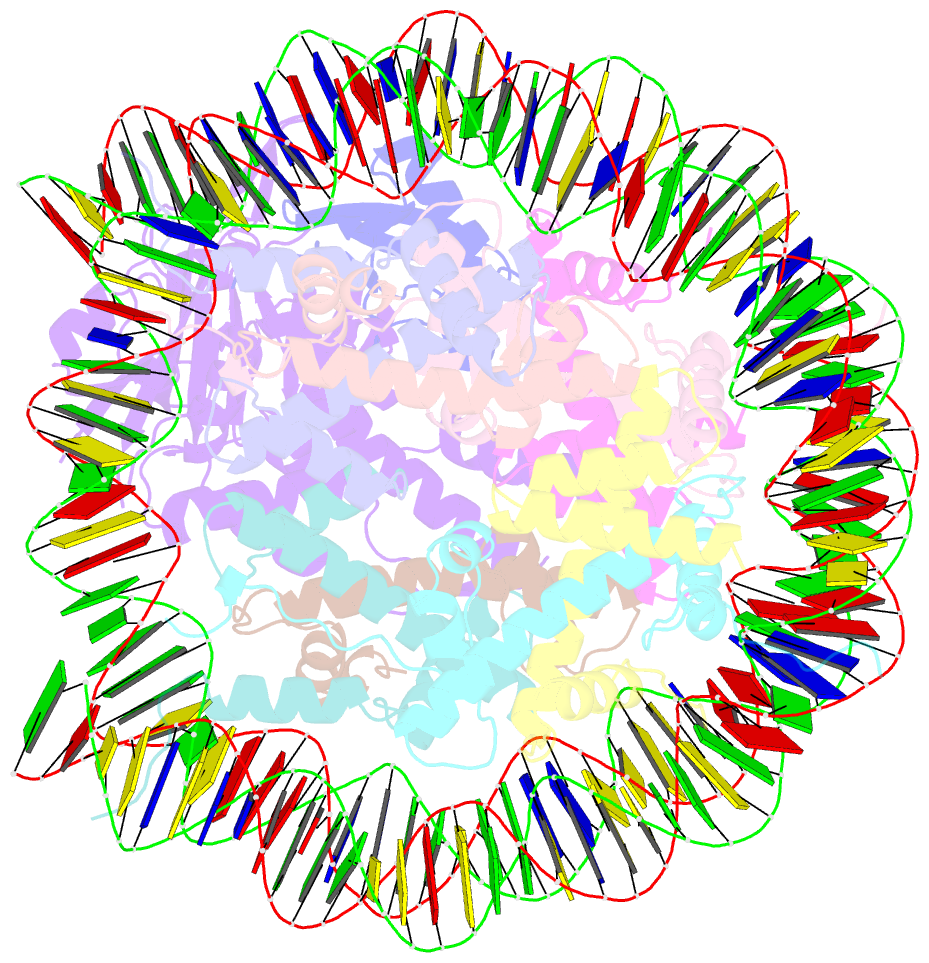
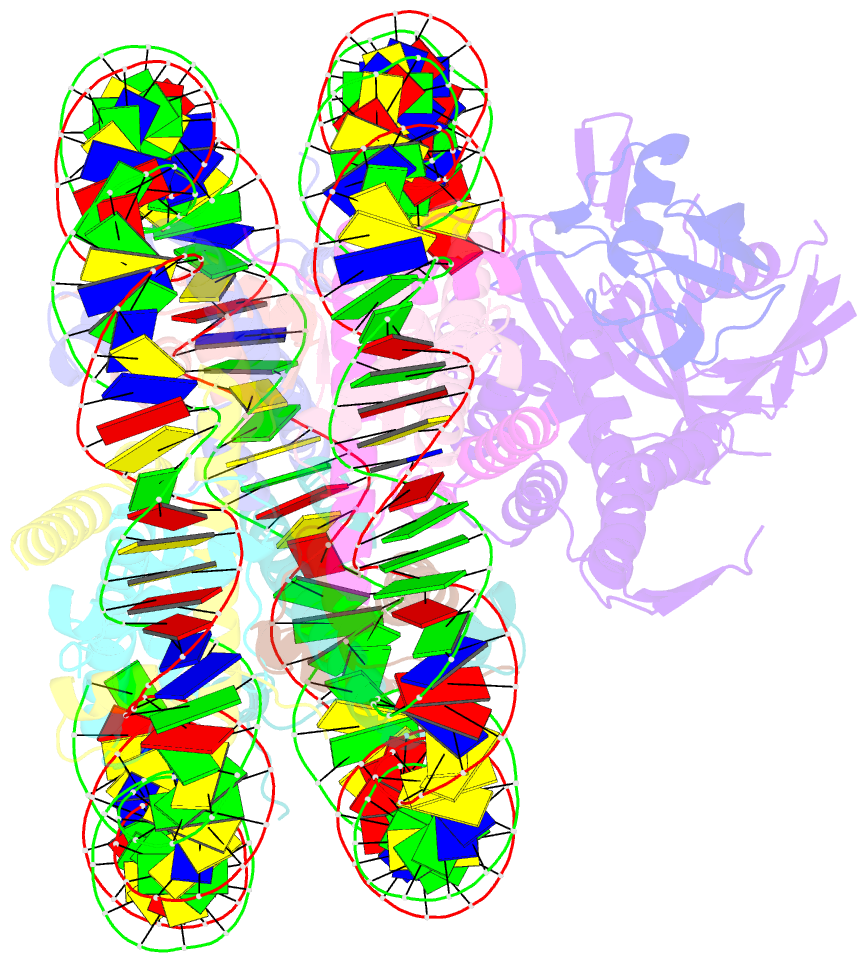
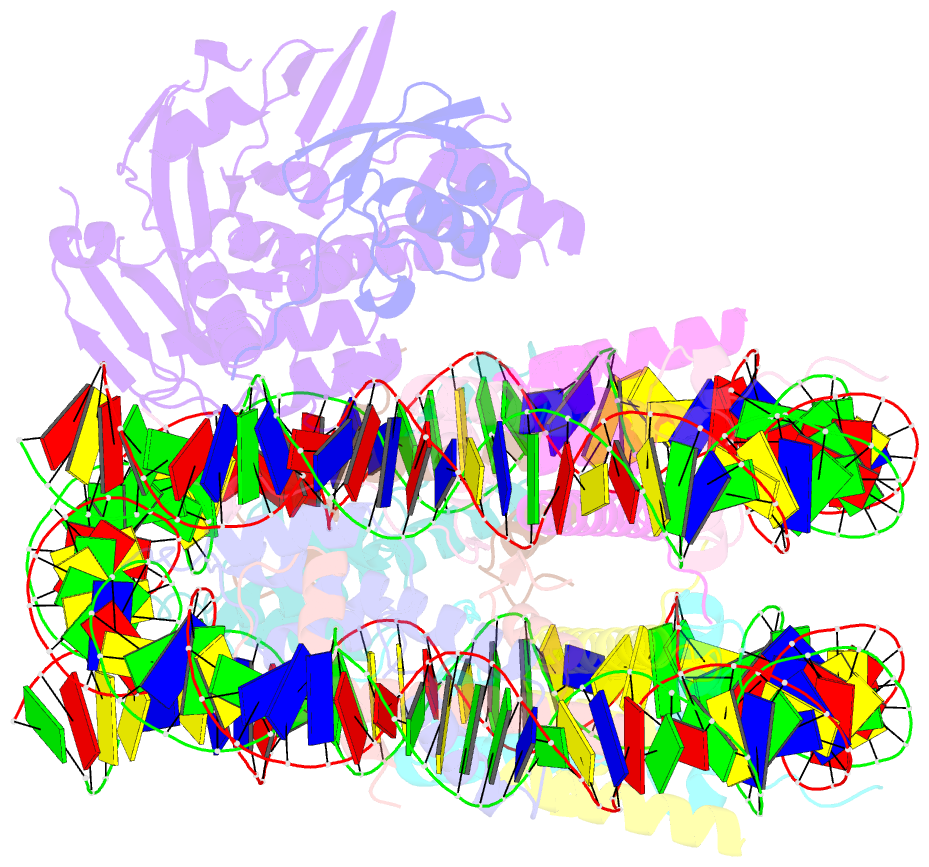
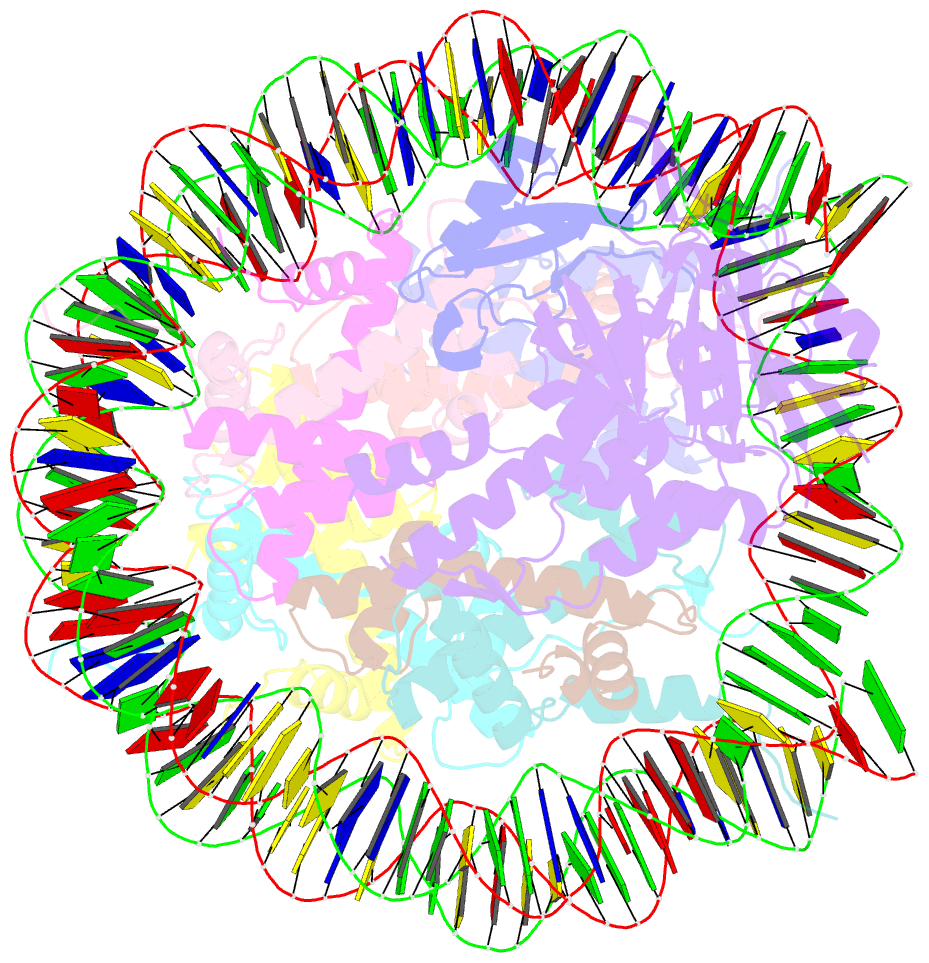
- cryo-EM structure of usp16 bound to h2ak119ub nucleosome
- Reference
- Ai H, He Z, Deng Z, Chu GC, Shi Q, Tong Z, Li JB, Pan M, Liu L (2024): "Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16." Nat.Struct.Mol.Biol. doi: 10.1038/s41594-024-01342-2.
- Abstract
- Epigenetic regulators have a crucial effect on gene expression based on their manipulation of histone modifications. Histone H2AK119 monoubiquitination (H2AK119Ub), a well-established hallmark in transcription repression, is dynamically regulated by the opposing activities of Polycomb repressive complex 1 (PRC1) and nucleosome deubiquitinases including the primary human USP16 and Polycomb repressive deubiquitinase (PR-DUB) complex. Recently, the catalytic mechanism for the multi-subunit PR-DUB complex has been described, but how the single-subunit USP16 recognizes the H2AK119Ub nucleosome and cleaves the ubiquitin (Ub) remains unknown. Here we report the cryo-EM structure of USP16-H2AK119Ub nucleosome complex, which unveils a fundamentally distinct mode of H2AK119Ub deubiquitination compared to PR-DUB, encompassing the nucleosome recognition pattern independent of the H2A-H2B acidic patch and the conformational heterogeneity in the Ub motif and the histone H2A C-terminal tail. Our work highlights the mechanism diversity of H2AK119Ub deubiquitination and provides a structural framework for understanding the disease-causing mutations of USP16.