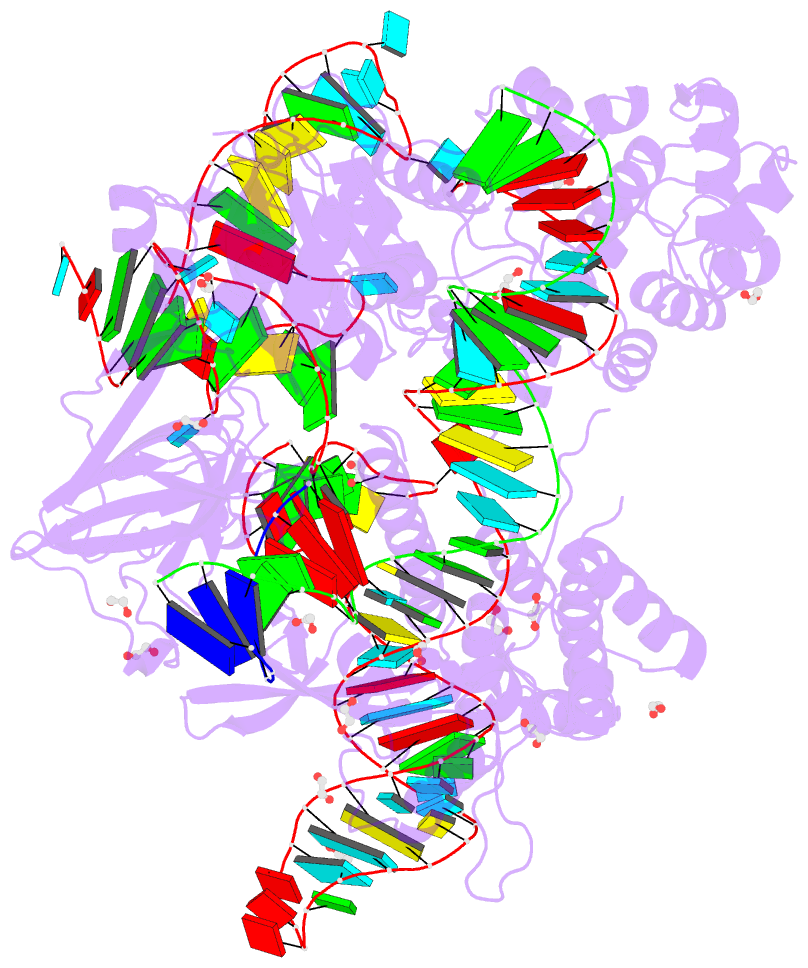
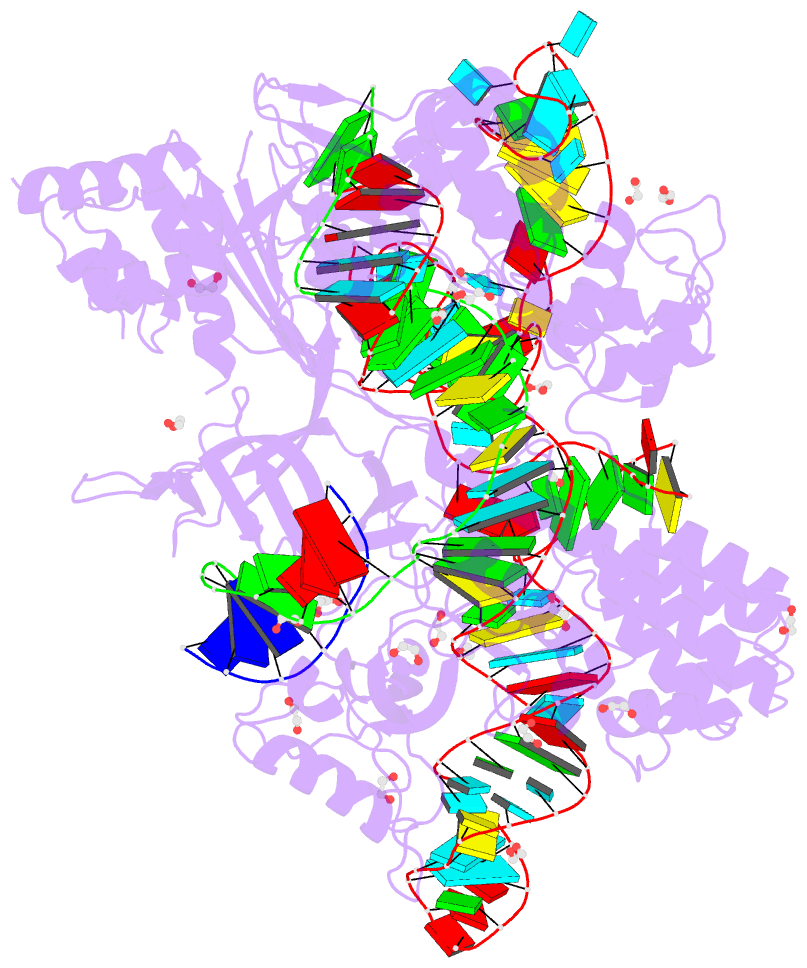
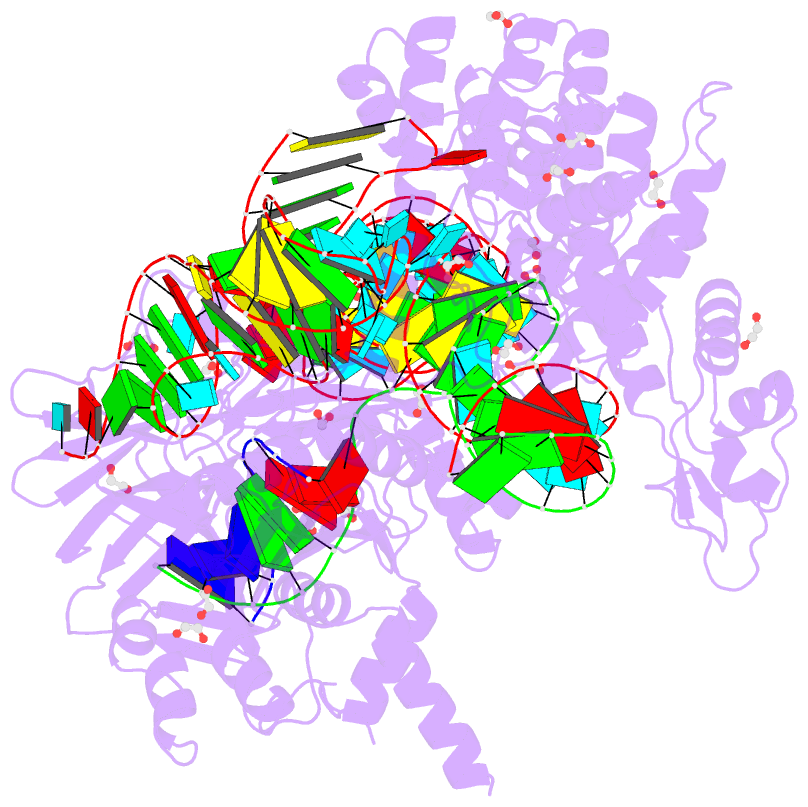
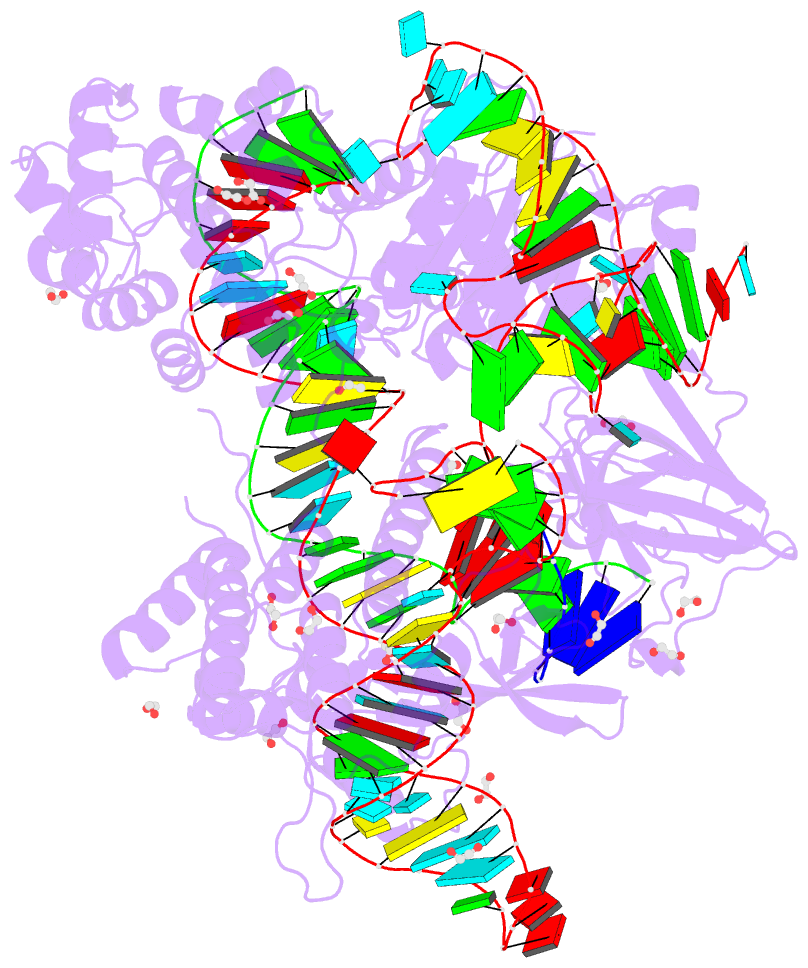
Summary information and primary citation
- PDB-id
- 8x5v; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (2.0 Å)
- Summary
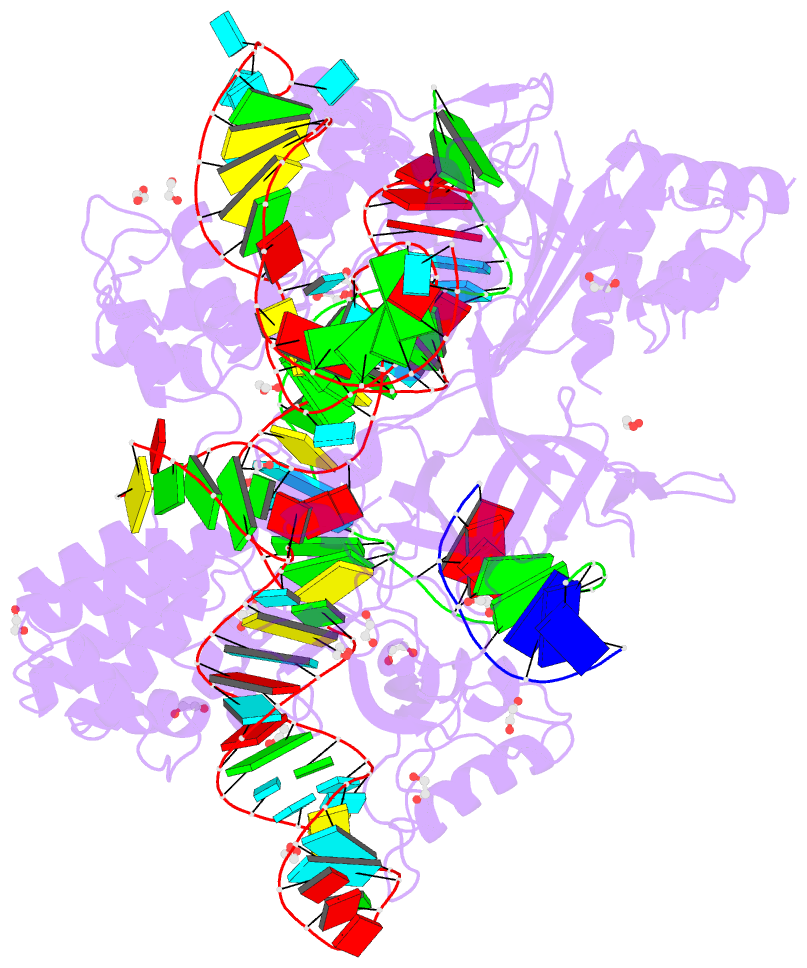
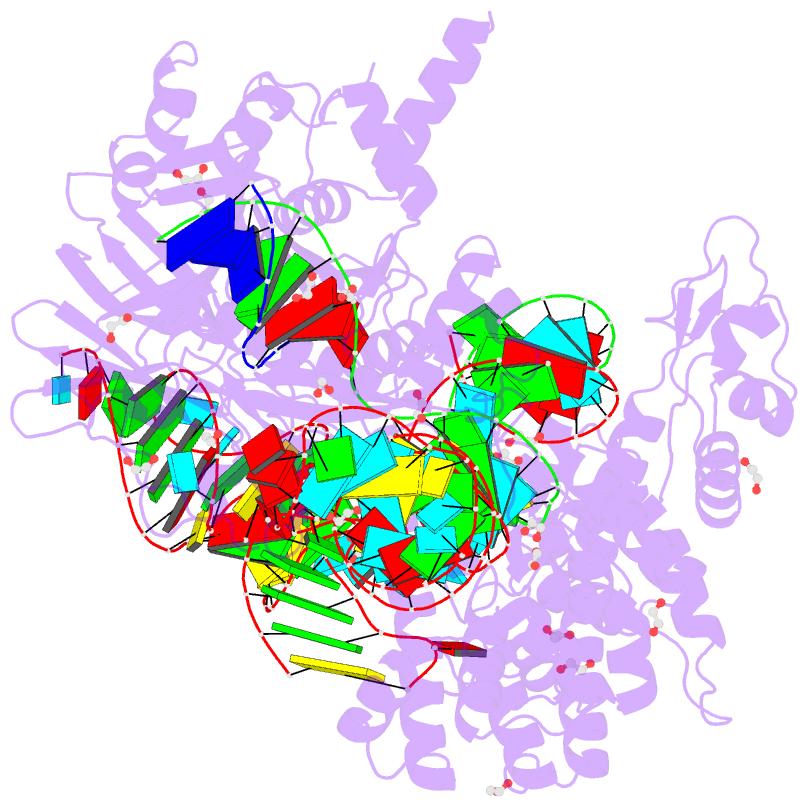
- Blcas9-sgrna-target DNA complex
- Reference
- Nakane T, Nakagawa R, Ishiguro S, Okazaki S, Mori H, Shuto Y, Yamashita K, Yachie N, Nishimasu H, Nureki O (2024): "Structure and engineering of Brevibacillus laterosporus Cas9." Commun Biol, 7, 803. doi: 10.1038/s42003-024-06422-z.
- Abstract
- The RNA-guided DNA endonuclease Cas9 cleaves double-stranded DNA targets complementary to an RNA guide, and is widely used as a powerful genome-editing tool. Here, we report the crystal structure of Brevibacillus laterosporus Cas9 (BlCas9, also known as BlatCas9), in complex with a guide RNA and its target DNA at 2.4-Å resolution. The structure reveals that the BlCas9 guide RNA adopts an unexpected architecture containing a triple-helix, which is specifically recognized by BlCas9, and that BlCas9 recognizes a unique N4CNDN protospacer adjacent motif through base-specific interactions on both the target and non-target DNA strands. Based on the structure, we rationally engineered a BlCas9 variant that exhibits enhanced genome- and base-editing activities with an expanded target scope in human cells. This approach may further improve the performance of the enhanced BlCas9 variant to generate useful genome-editing tools that require only a single C PAM nucleotide and can be packaged into a single AAV vector for in vivo gene therapy.