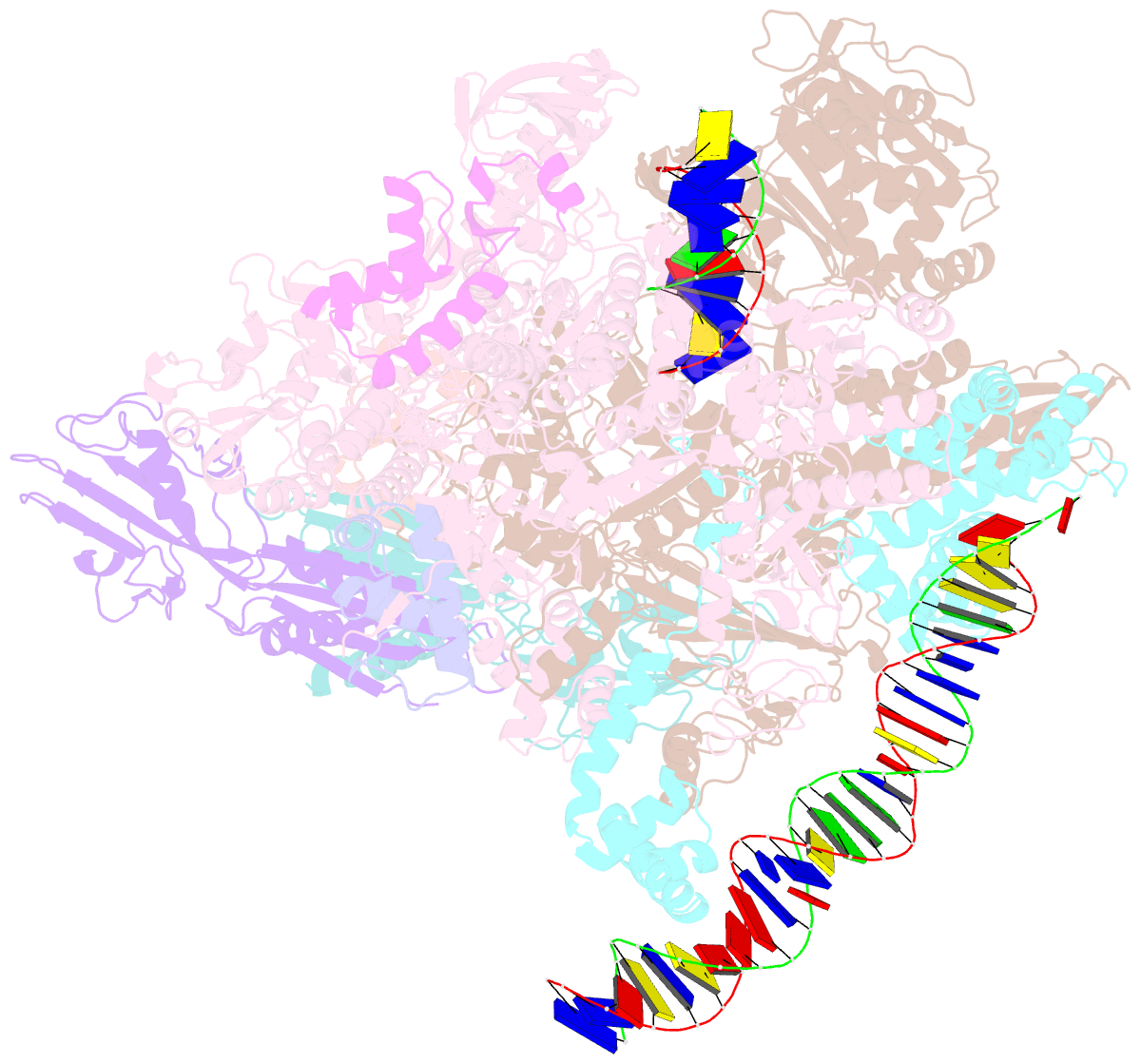
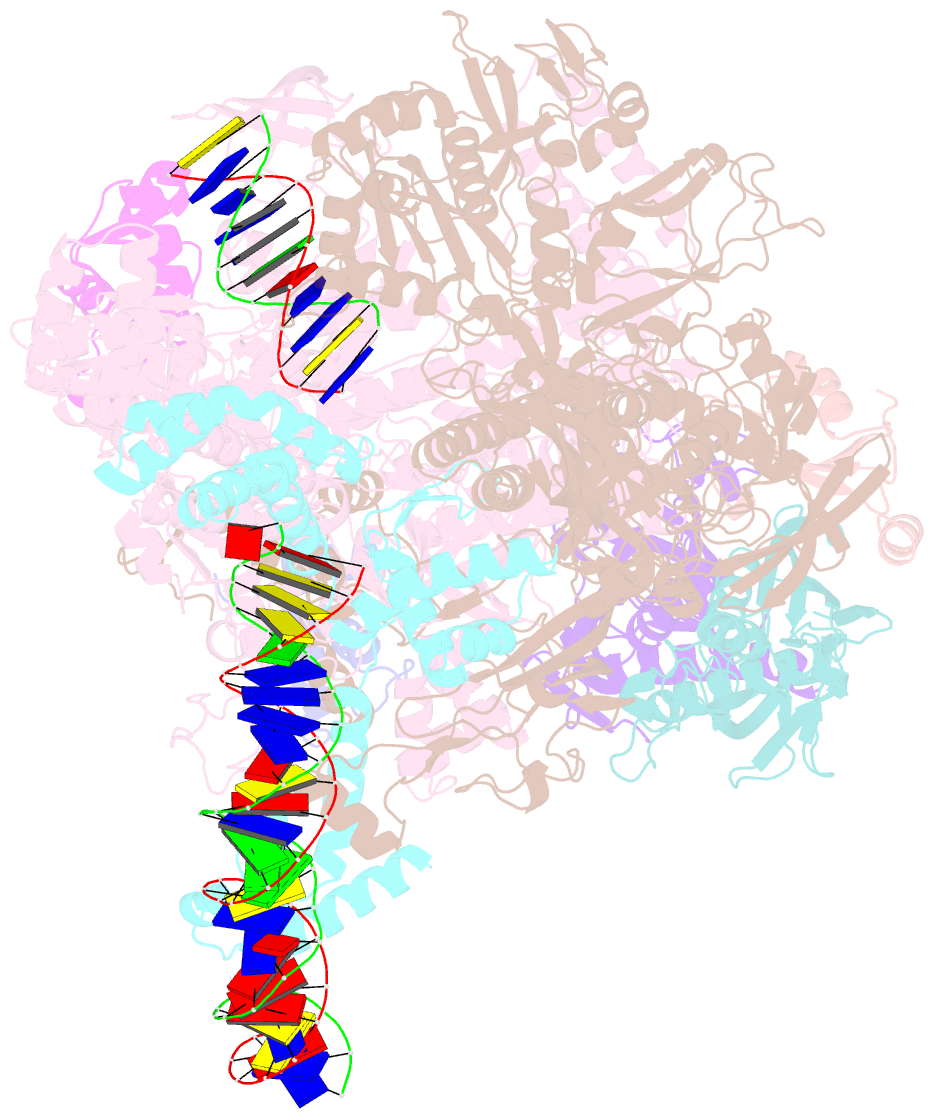
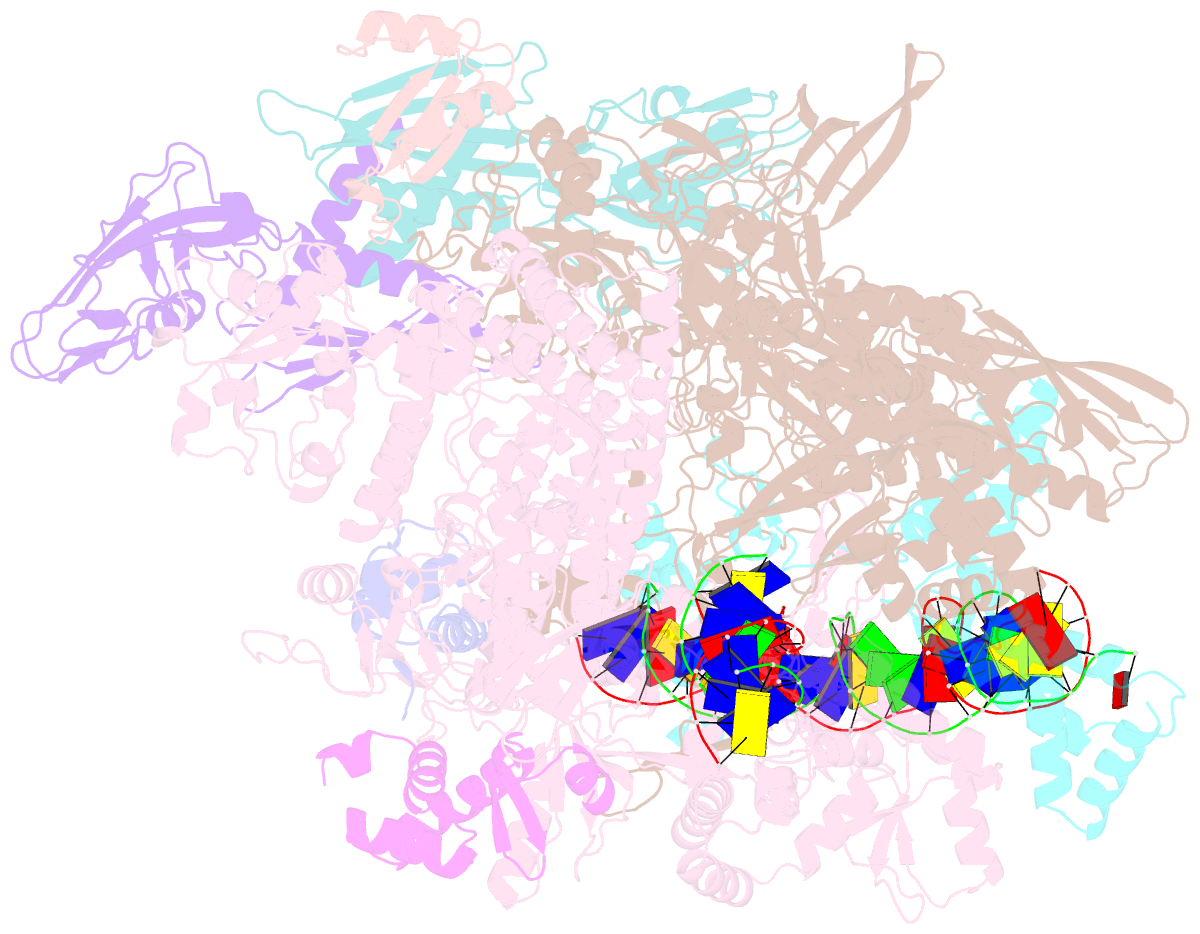
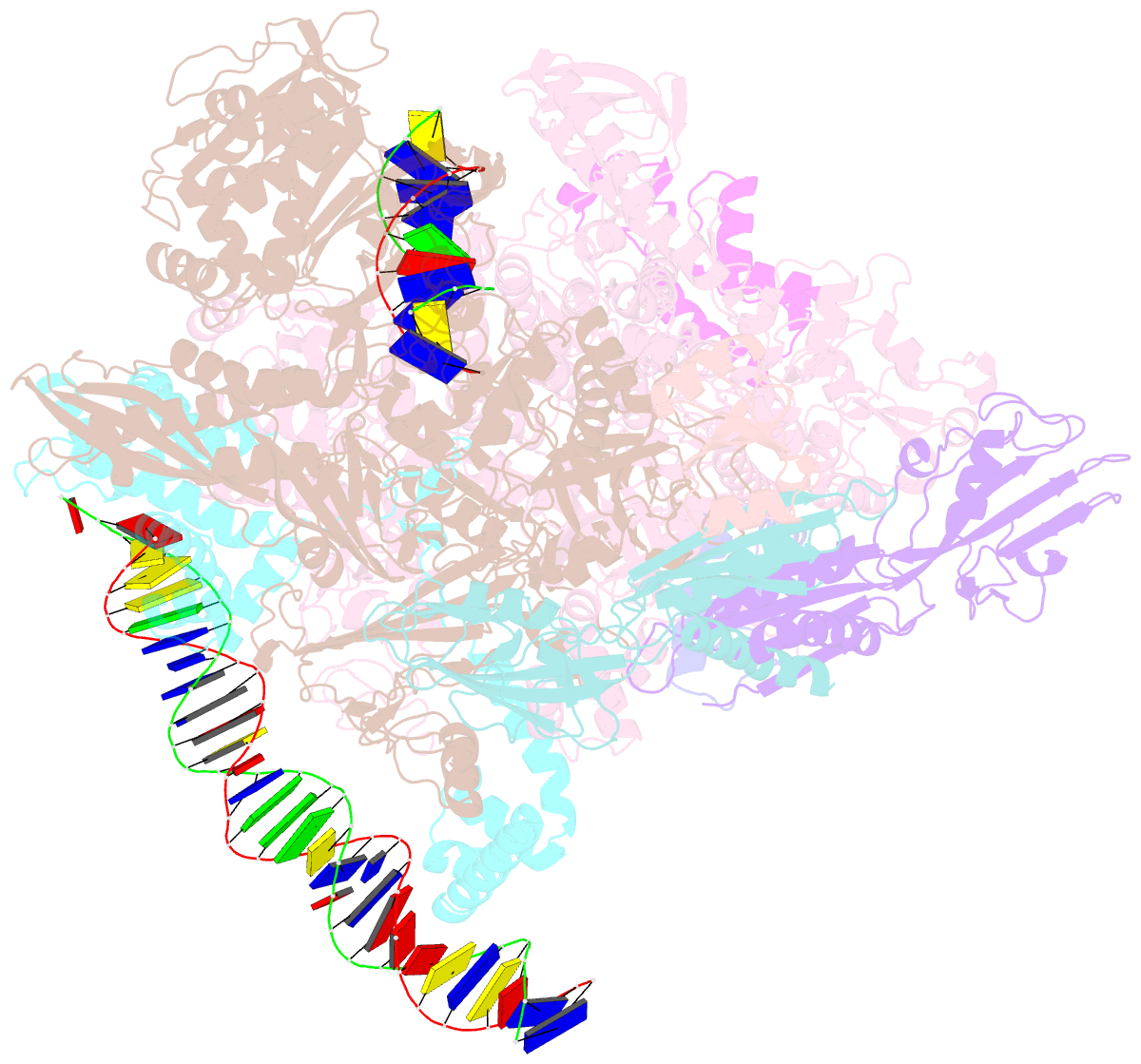
Summary information and primary citation
- PDB-id
- 8x6g; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.3 Å)
- Summary
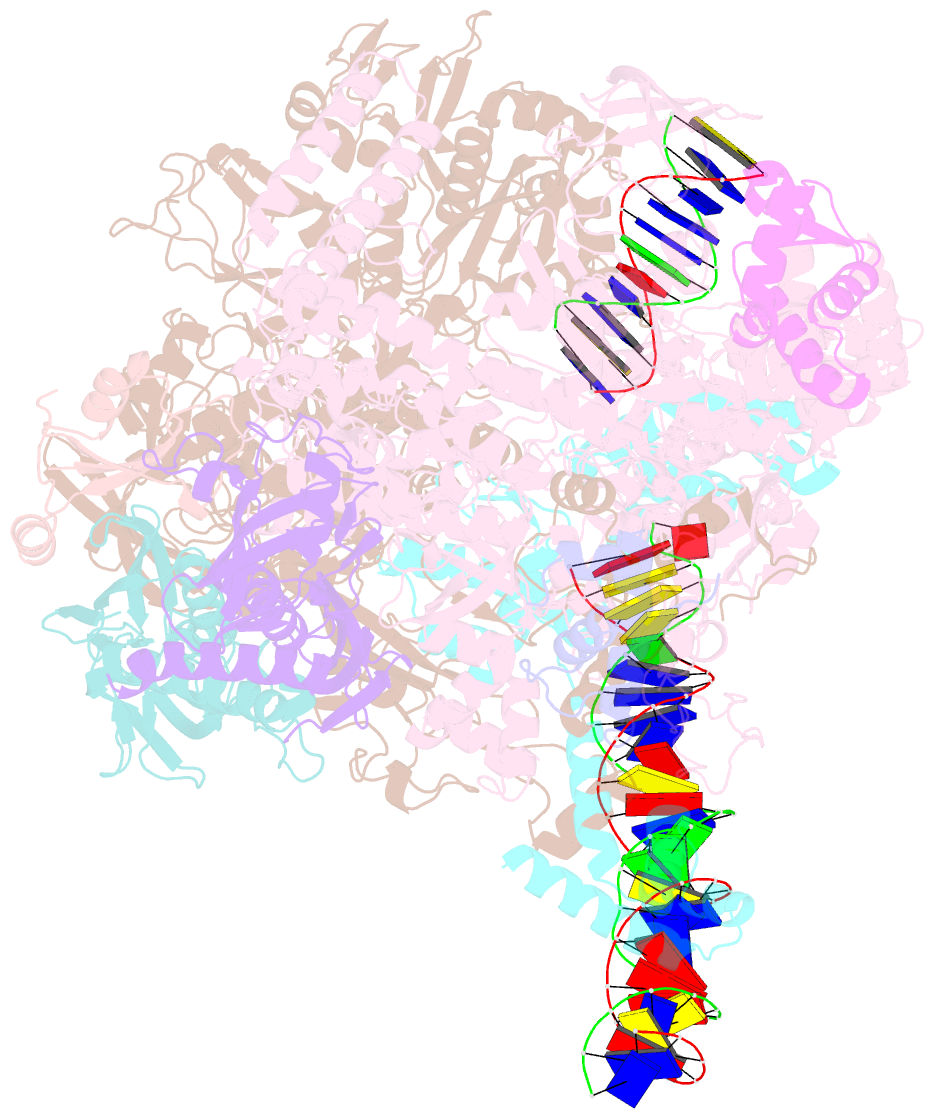
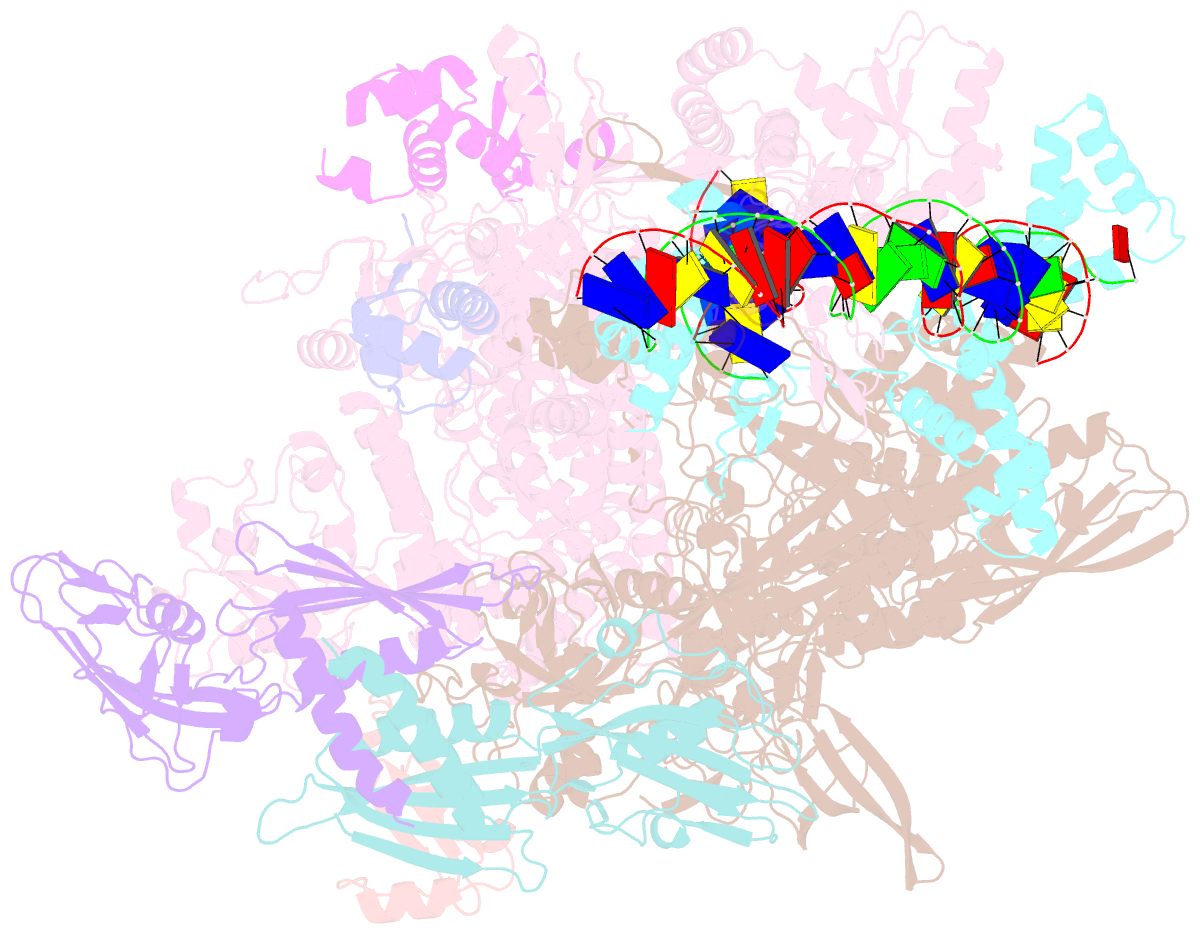
- cryo-EM structure of staphylococcus aureus sigb-dependent rnap-promoter open complex
- Reference
- Yuan L, Liu Q, Xu L, Wu B, Feng Y (2024): "Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase." Nat Commun, 15, 4850. doi: 10.1038/s41467-024-49229-6.
- Abstract
- Bacterial RNAP needs to form holoenzyme with σ factors to initiate transcription. While Staphylococcus aureus σA controls housekeeping functions, S. aureus σB regulates virulence, biofilm formation, persistence, cell internalization, membrane transport, and antimicrobial resistance. Besides the sequence difference, the spacers between the -35 element and -10 element of σB regulated promoters are shorter than those of σA regulated promoters. Therefore, how σB recognizes and initiates transcription from target promoters can not be inferred from that of the well studied σ. Here, we report the cryo-EM structures of S. aureus RNAP-promoter open complexes comprising σA and σB, respectively. Structural analyses, in combination with biochemical experiments, reveal the structural basis for the promoter specificity of S. aureus transcription. Although the -10 element of σA regulated promoters is recognized by domain σA2 as single-stranded DNA, the -10 element of σB regulated promoters is co-recognized by domains σB2 and σB3 as double-stranded DNA, accounting for the short spacers of σB regulated promoters. S. aureus RNAP is a validated target of antibiotics, and our structures pave the way for rational drug design targeting S. aureus RNAP.