Summary information and primary citation
- PDB-id
- 8y7g; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (3.15 Å)
- Summary
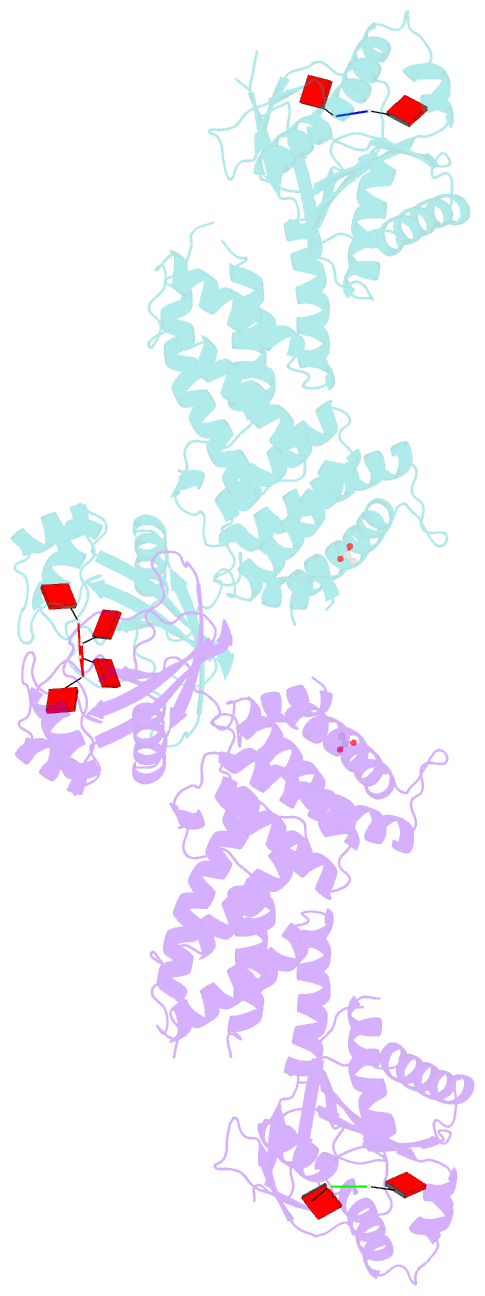
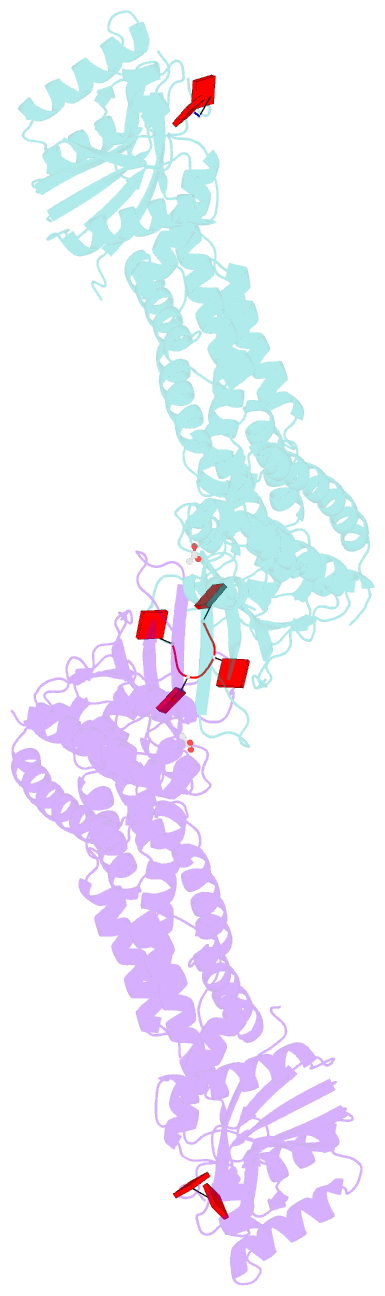
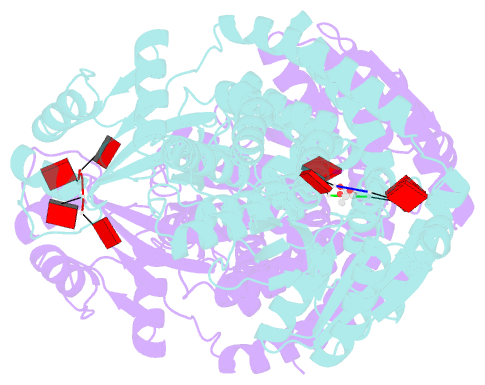
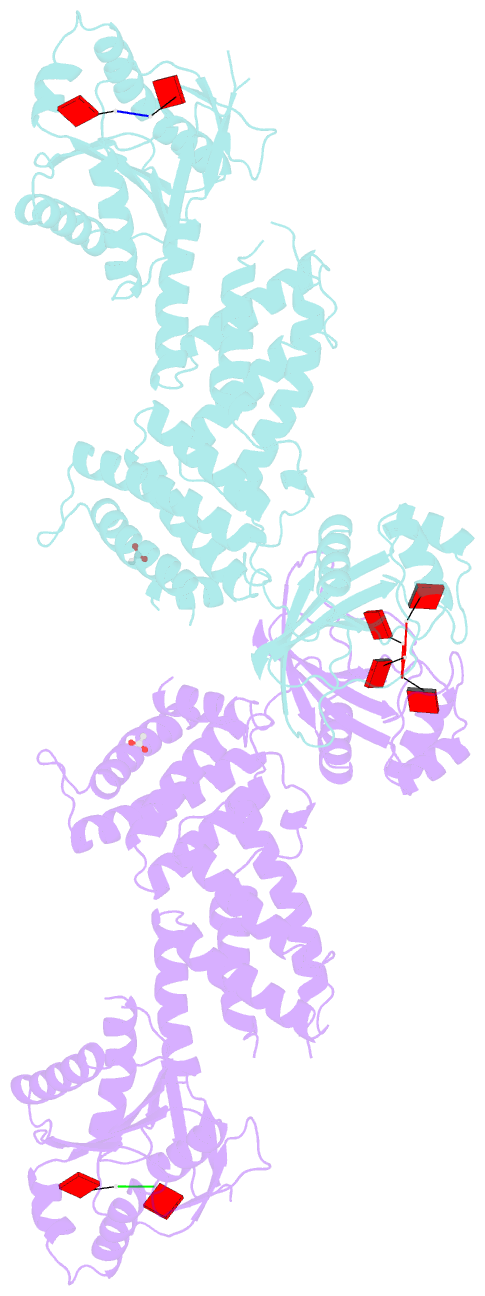
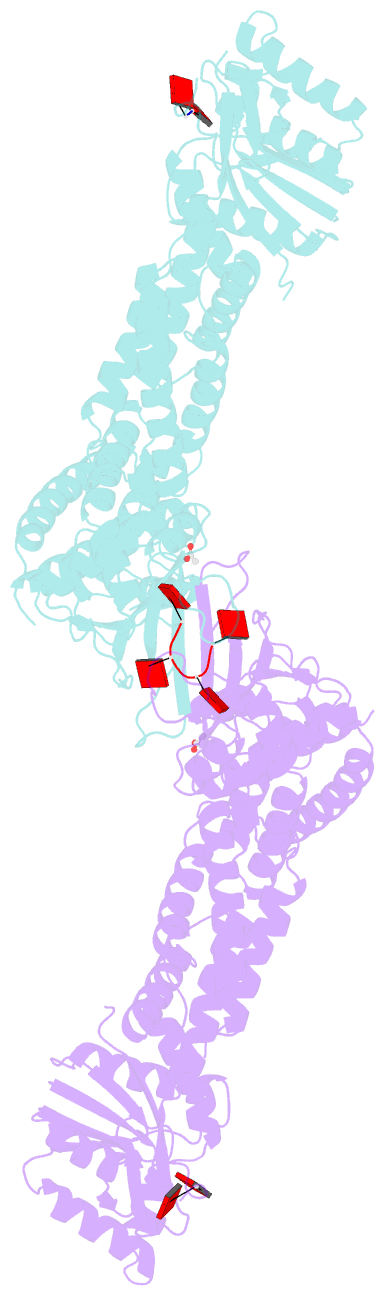
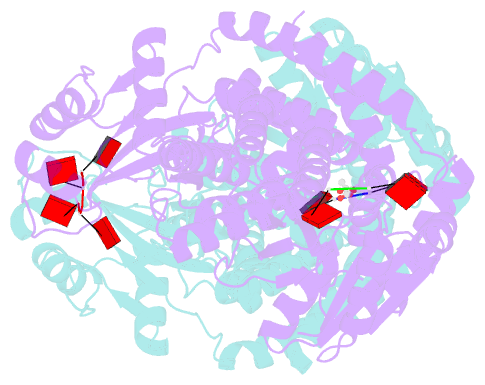
- Crystal structure of the marinitoga sp. csx1-crn2 h495a mutant in complex with cyclic-tetraadenylate (ca4)
- Reference
- Zhang D, Du L, Gao H, Yuan C, Lin Z (2024): "Structural insight into the Csx1-Crn2 fusion self-limiting ribonuclease of type III CRISPR system." Nucleic Acids Res., 52, 8419-8430. doi: 10.1093/nar/gkae569.
- Abstract
- In the type III CRISPR system, cyclic oligoadenylate (cOA) molecules act as second messengers, activating various promiscuous ancillary nucleases that indiscriminately degrade host and viral DNA/RNA. Conversely, ring nucleases, by specifically cleaving cOA molecules, function as off-switches to protect host cells from dormancy or death, and allow viruses to counteract immune responses. The fusion protein Csx1-Crn2, combining host ribonuclease with viral ring nuclease, represents a unique self-limiting ribonuclease family. Here, we describe the structures of Csx1-Crn2 from the organism of Marinitoga sp., in both its full-length and truncated forms, as well as in complex with cA4. We show that Csx1-Crn2 operates as a homo-tetramer, a configuration crucial for preserving the structural integrity of the HEPN domain and ensuring effective ssRNA cleavage. The binding of cA4 to the CARF domain triggers significant conformational changes across the CARF, HTH, and into the HEPN domains, leading the two R-X4-6-H motifs to form a composite catalytic site. Intriguingly, an acetate ion was found to bind at this composite site by mimicking the scissile phosphate. Further molecular docking analysis reveals that the HEPN domain can accommodate a single ssRNA molecule involving both R-X4-6-H motifs, underscoring the importance of HEPN domain dimerization for its activation.