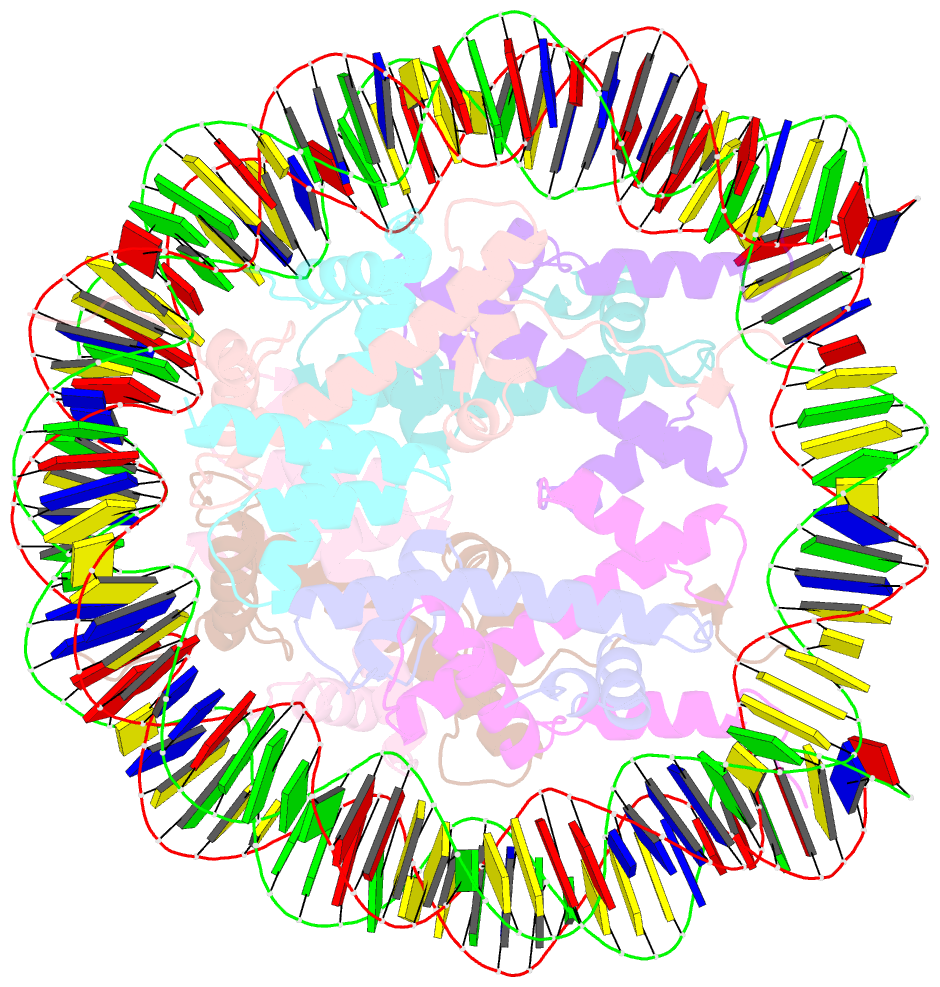
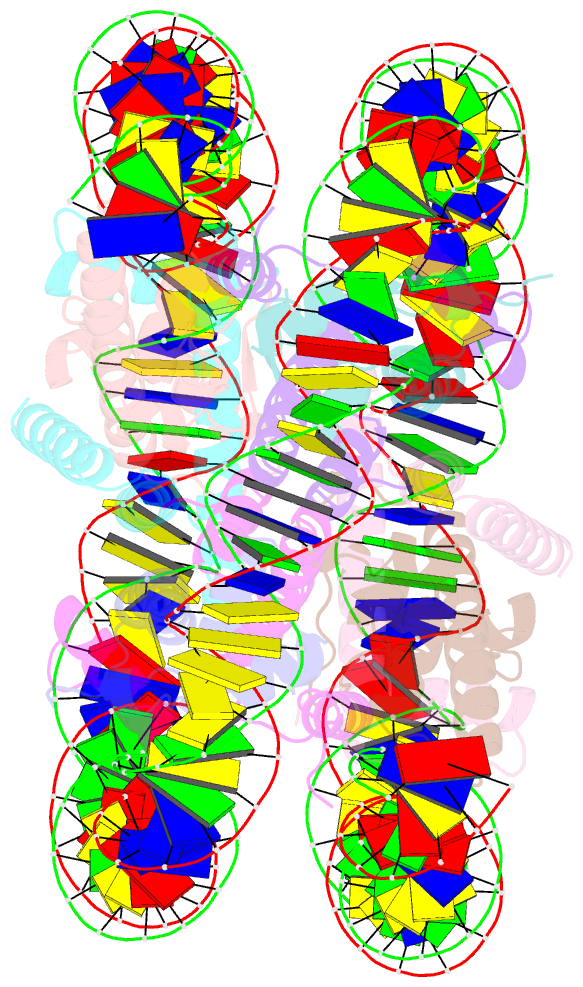
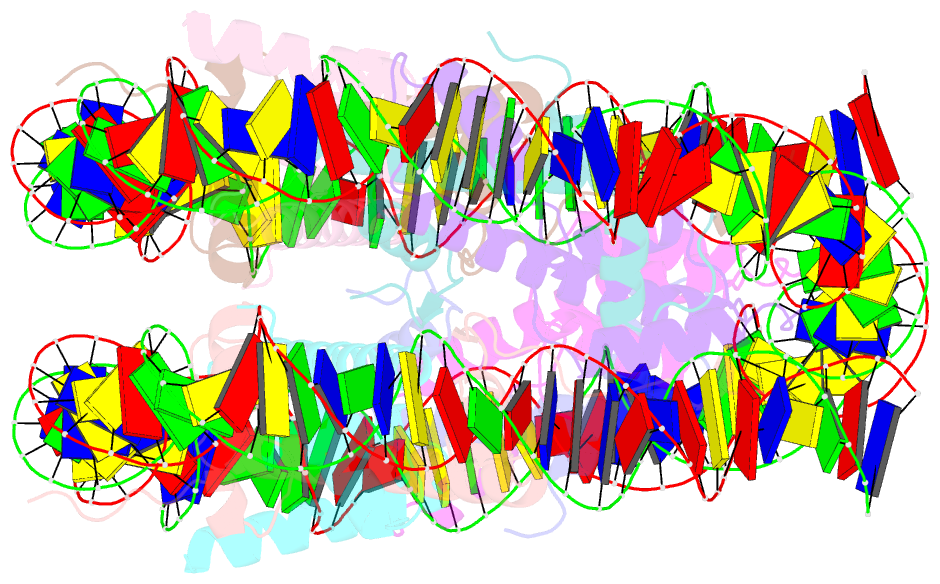
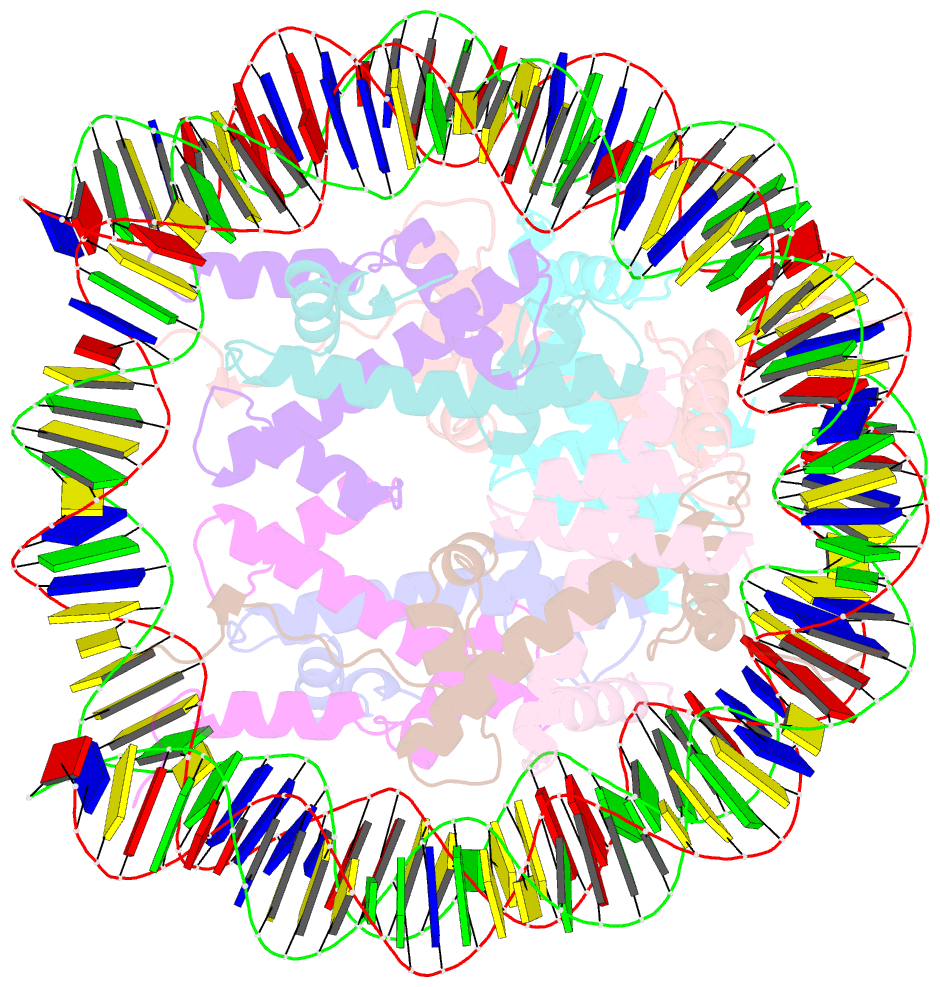
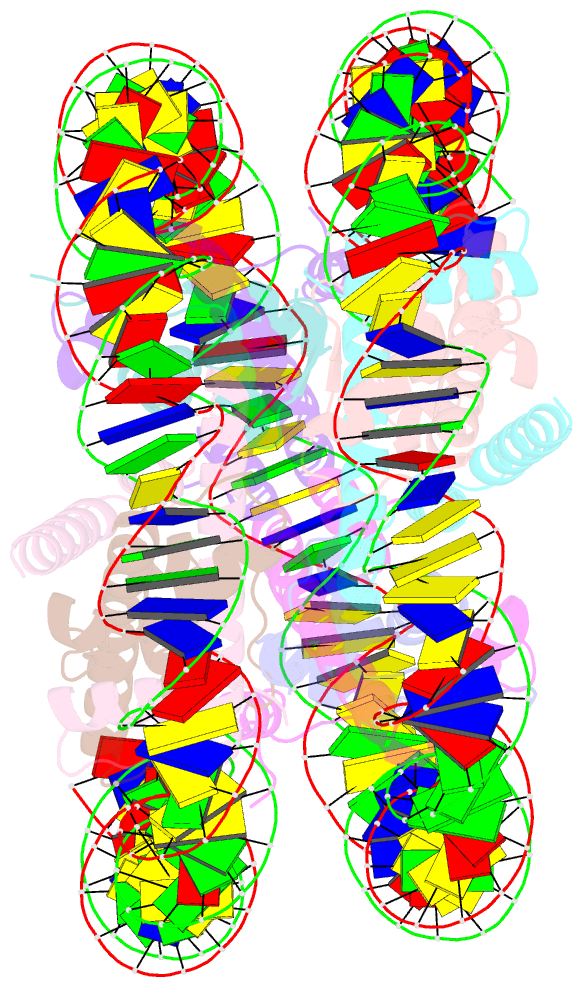
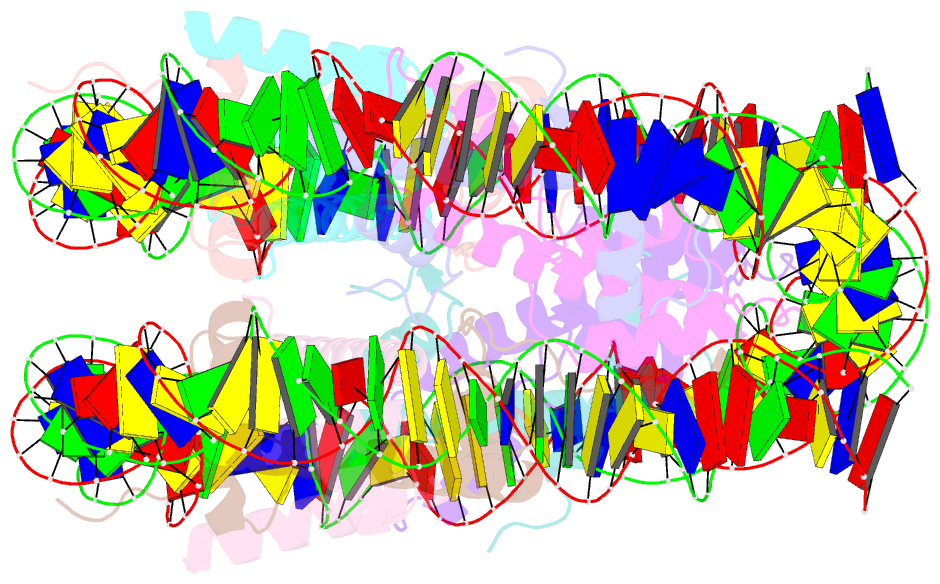
Summary information and primary citation
- PDB-id
- 8ybj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (2.38 Å)
- Summary
- cryo-EM structure of human nucleosome core particle composed of the widom 601 DNA sequence
- Reference
- Kimura T, Hirai S, Kujirai T, Fujita R, Ogasawara M, Ehara H, Sekine SI, Takizawa Y, Kurumizaka H (2024): "Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K." Genes Cells, 29, 769-781. doi: 10.1111/gtc.13143.
- Abstract
- The Lys mutation of the canonical histone H3.1 Glu97 residue (H3E97K) is found in cancer cells. Previous biochemical analyses revealed that the nucleosome containing the H3E97K mutation is extremely unstable as compared to the wild-type nucleosome. However, the mechanism by which the H3E97K mutation causes nucleosome instability has not been clarified yet. In the present study, the cryo-electron microscopy structure of the nucleosome containing the H3E97K mutation revealed that the entry/exit DNA regions of the H3E97K nucleosome are disordered, probably by detachment of the nucleosomal DNA from the H3 N-terminal regions. This may change the intra-molecular amino acid interactions with the replaced H3 Lys97 residue, inducing structural distortion around the mutated position in the nucleosome. Consistent with the nucleosomal DNA end flexibility and the nucleosome instability, the H3E97K mutation exhibited reduced binding of linker histone H1 to the nucleosome, defective activation of PRC2 (the essential methyltransferase for facultative heterochromatin formation) with a poly-nucleosome, and enhanced nucleosome transcription by RNA polymerase II.