Summary information and primary citation
- PDB-id
- 8yfq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (3.3 Å)
- Summary
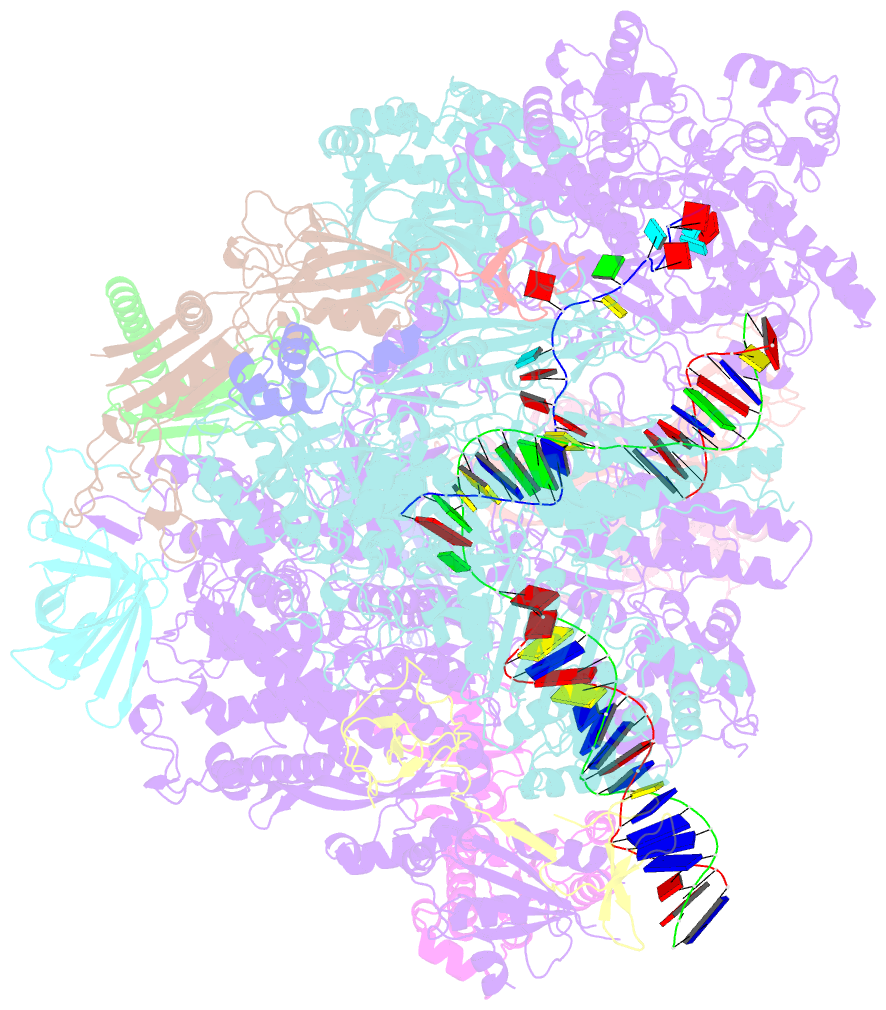
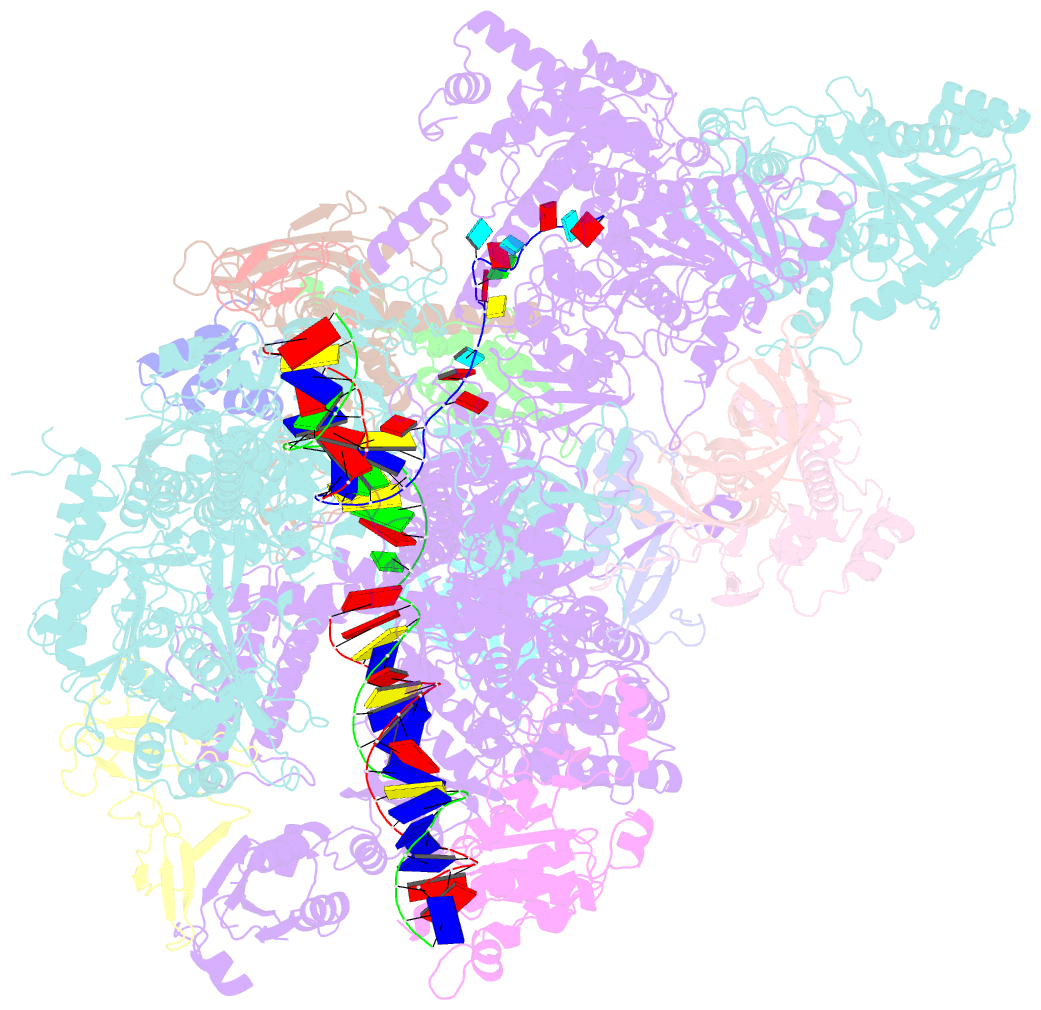
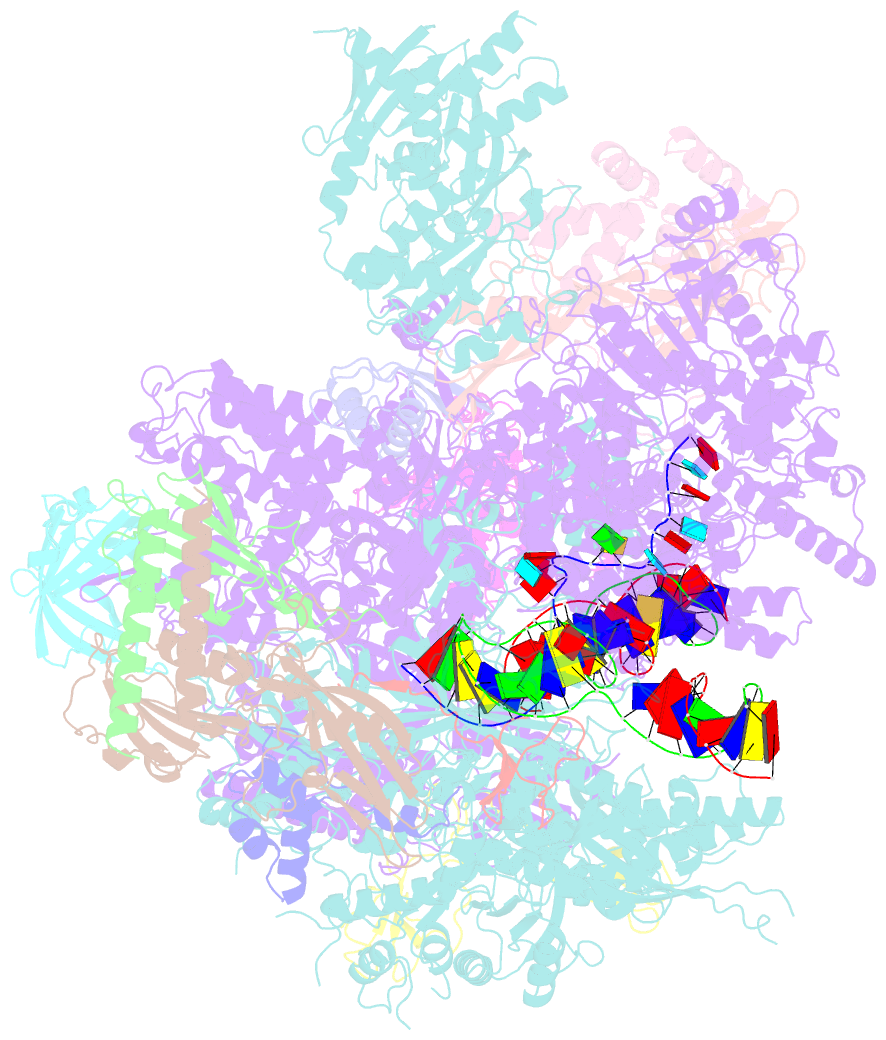
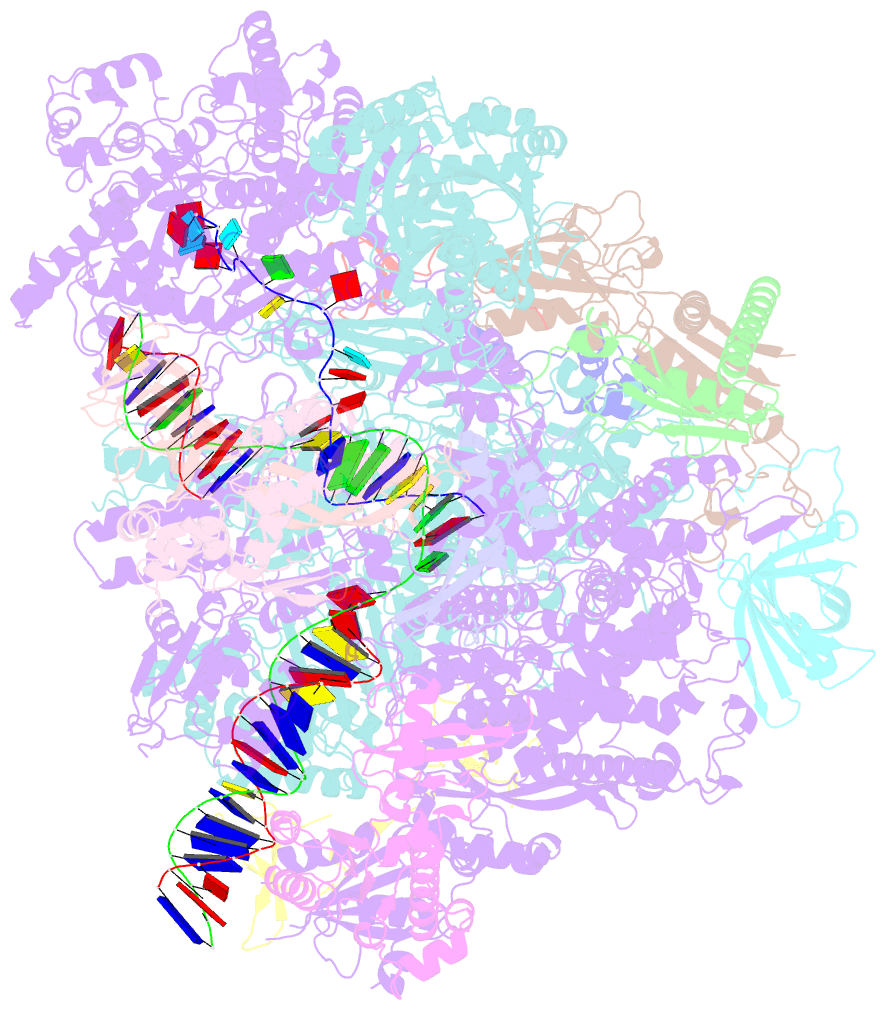
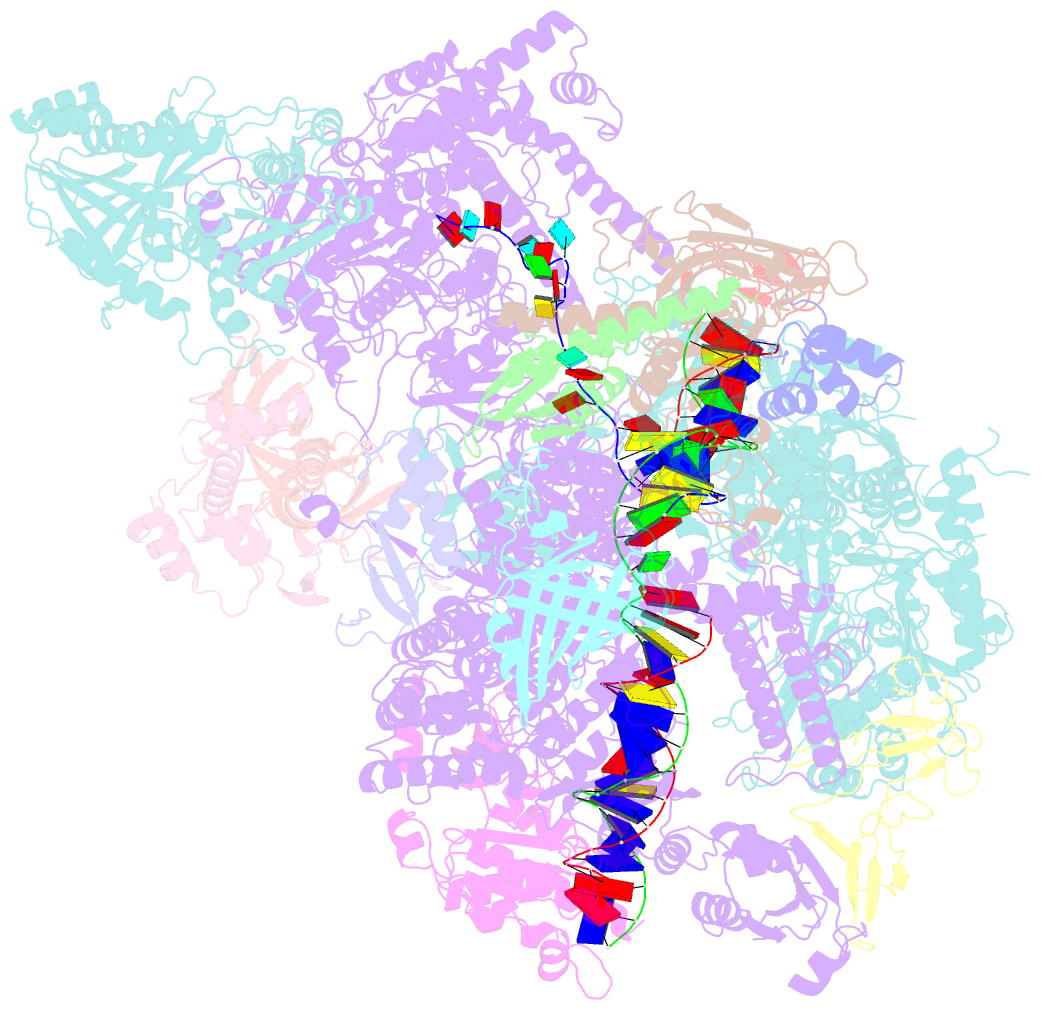
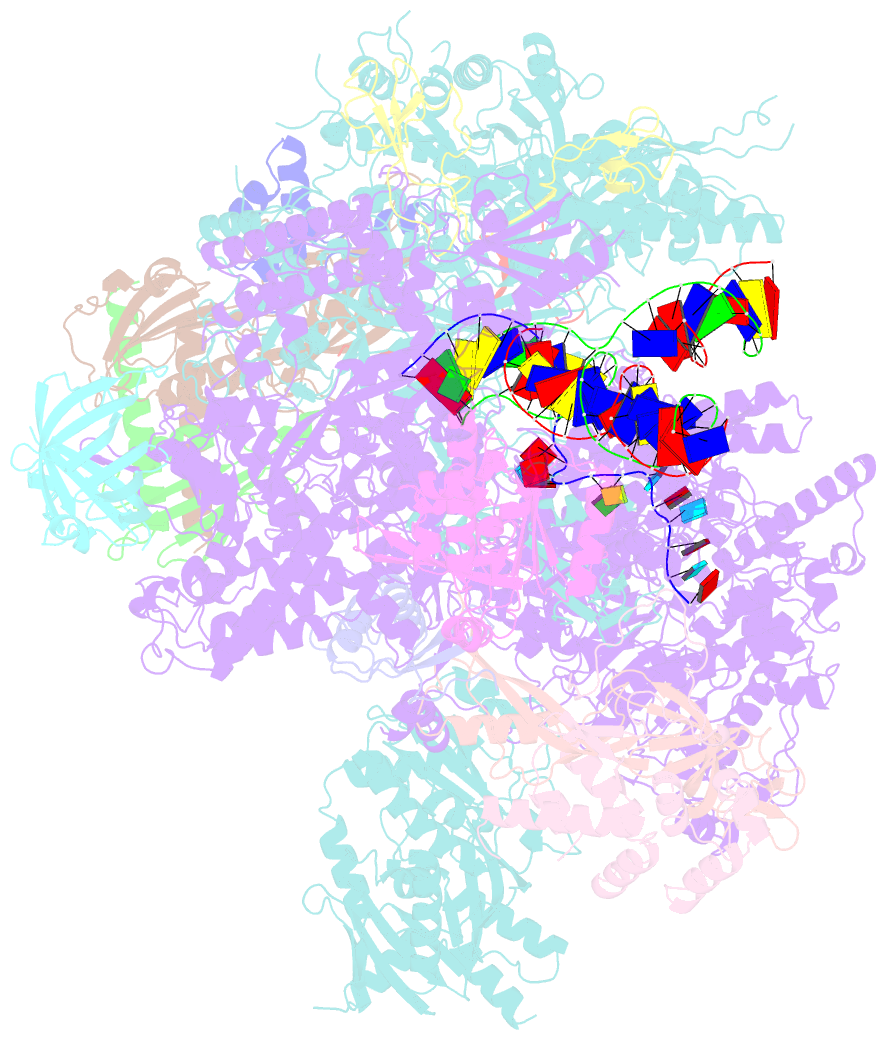
- Cryo em structure of komagataella phaffii rnapii-rat1-rai1 pre-termination complex
- Reference
- Yanagisawa T, Murayama Y, Ehara H, Goto M, Aoki M, Sekine SI (2024): "Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex." Nat Commun, 15, 7854. doi: 10.1038/s41467-024-52157-0.
- Abstract
- The 5´-3´ exoribonuclease Rat1/Xrn2 is responsible for the termination of eukaryotic mRNA transcription by RNAPII. Rat1 forms a complex with its partner proteins, Rai1 and Rtt103, and acts as a "torpedo" to bind transcribing RNAPII and dissociate DNA/RNA from it. Here we report the cryo-electron microscopy structures of the Rat1-Rai1-Rtt103 complex and three Rat1-Rai1-associated RNAPII complexes (type-1, type-1b, and type-2) from the yeast, Komagataella phaffii. The Rat1-Rai1-Rtt103 structure revealed that Rat1 and Rai1 form a heterotetramer with a single Rtt103 bound between two Rai1 molecules. In the type-1 complex, Rat1-Rai1 forms a heterodimer and binds to the RNA exit site of RNAPII to extract RNA into the Rat1 exonuclease active site. This interaction changes the RNA path in favor of termination (the "pre-termination" state). The type-1b and type-2 complexes have no bound DNA/RNA, likely representing the "post-termination" states. These structures illustrate the termination mechanism of eukaryotic mRNA transcription.