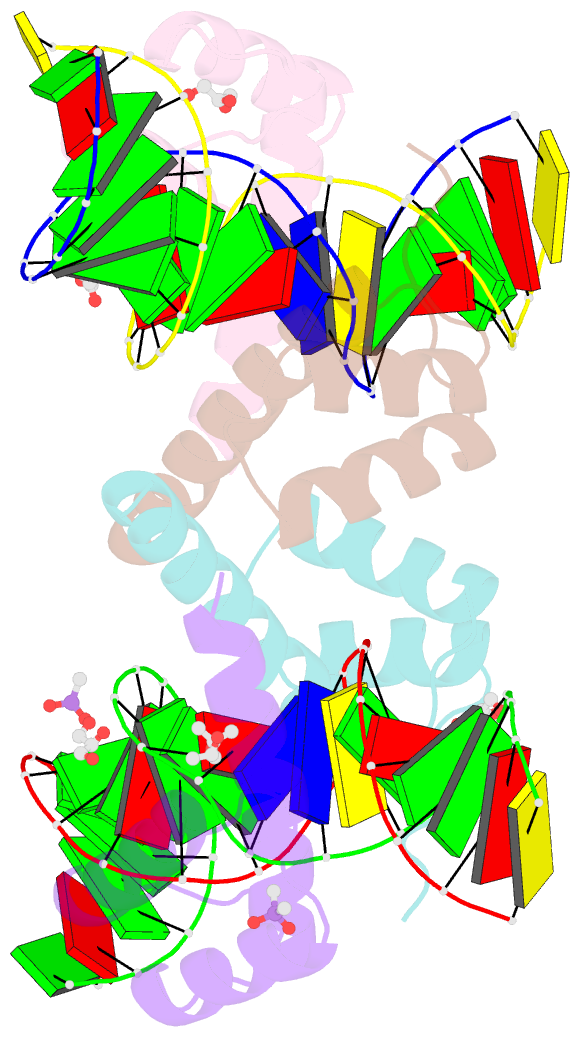
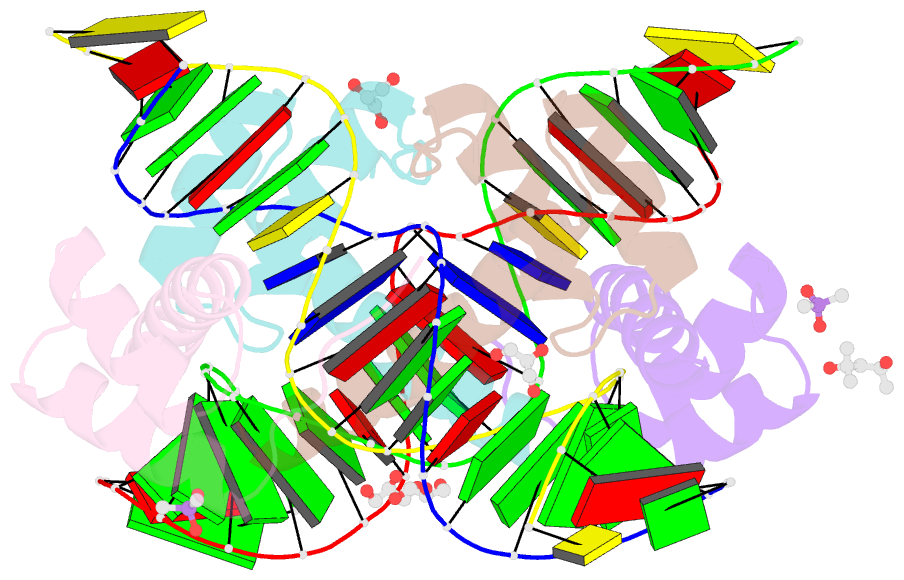
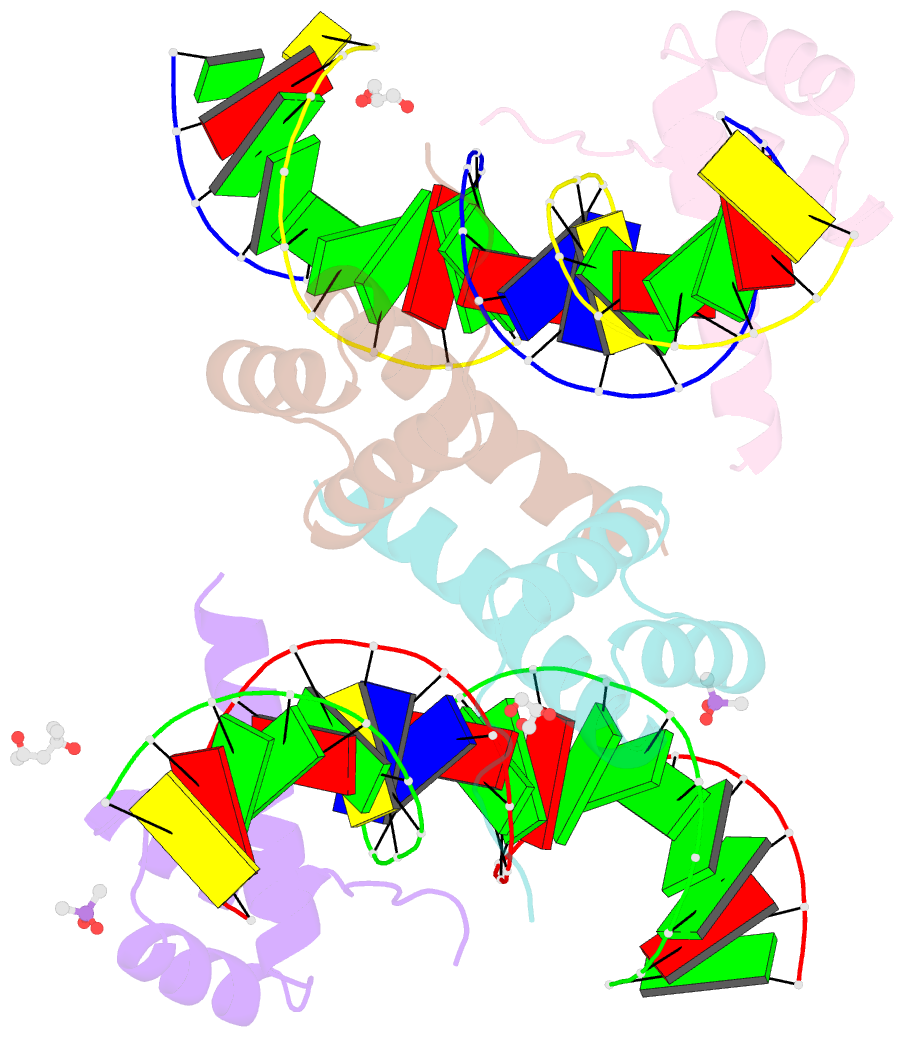
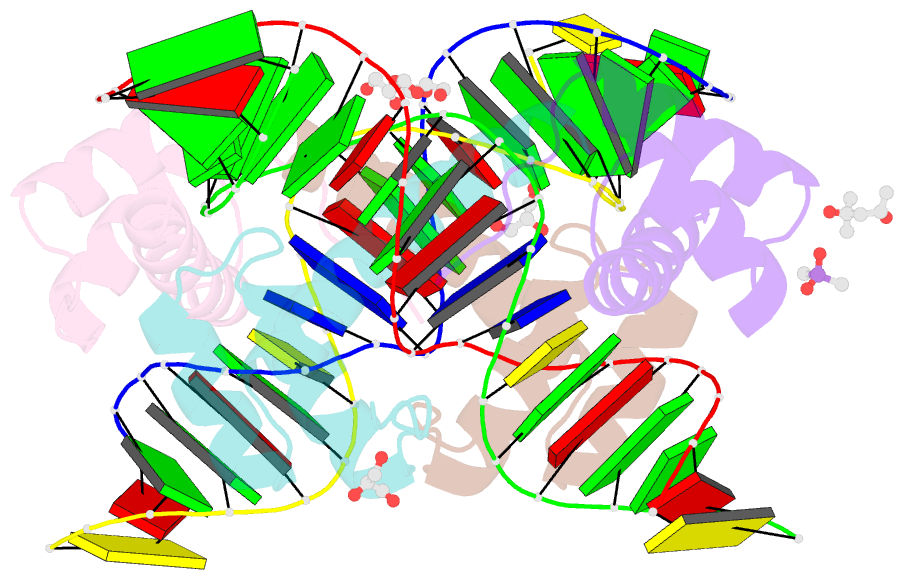
Summary information and primary citation
- PDB-id
- 9b8u; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.9 Å)
- Summary
- Crystal structure of crx-ret4 oligonucleotide complex
- Reference
- Srivastava D, Gowribidanur-Chinnaswamy P, Gaur P, Spies M, Swaroop A, Artemyev NO (2024): "Molecular basis of CRX/DNA recognition and stoichiometry at the Ret4 response element." Structure, 32, 1751-1759.e4. doi: 10.1016/j.str.2024.07.004.
- Abstract
- Two retinal transcription factors, cone-rod homeobox (CRX) and neural retina leucine zipper (NRL), cooperate functionally and physically to control photoreceptor development and homeostasis. Mutations in CRX and NRL cause severe retinal diseases. Despite the roles of NRL and CRX, insight into their functions at the molecular level is lacking. Here, we have solved the crystal structure of the CRX homeodomain in complex with its cognate response element (Ret4) from the rhodopsin proximal promoter region. The structure reveals an unexpected 2:1 stoichiometry of CRX/Ret4 and unique orientation of CRX molecules on DNA, and it explains the mechanisms of pathogenic mutations in CRX. Mutations R41Q and E42K disrupt the CRX protein-protein contacts based on the structure and reduce the CRX/Ret4 binding stoichiometry, suggesting a novel disease mechanism. Furthermore, we show that NRL alters the stoichiometry and increases affinity of CRX binding at the rhodopsin promoter, which may enhance transcription of rod-specific genes and suppress transcription of cone-specific genes.