Summary information and primary citation
- PDB-id
- 9bct; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- cryo-EM (2.5 Å)
- Summary
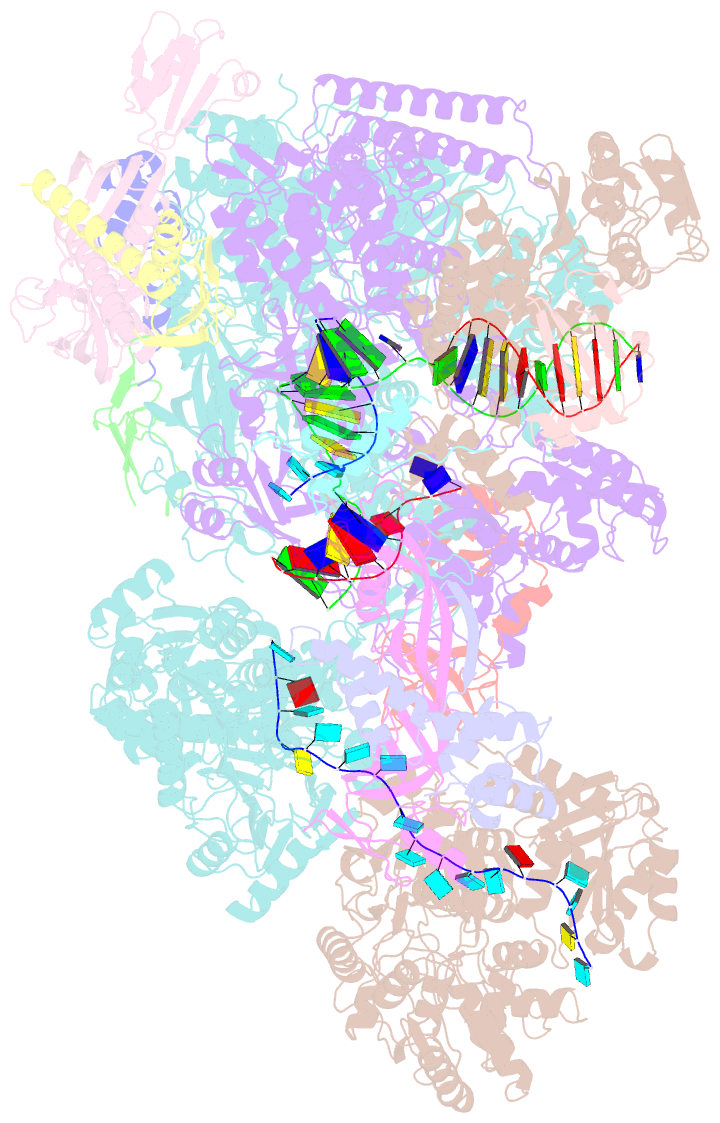
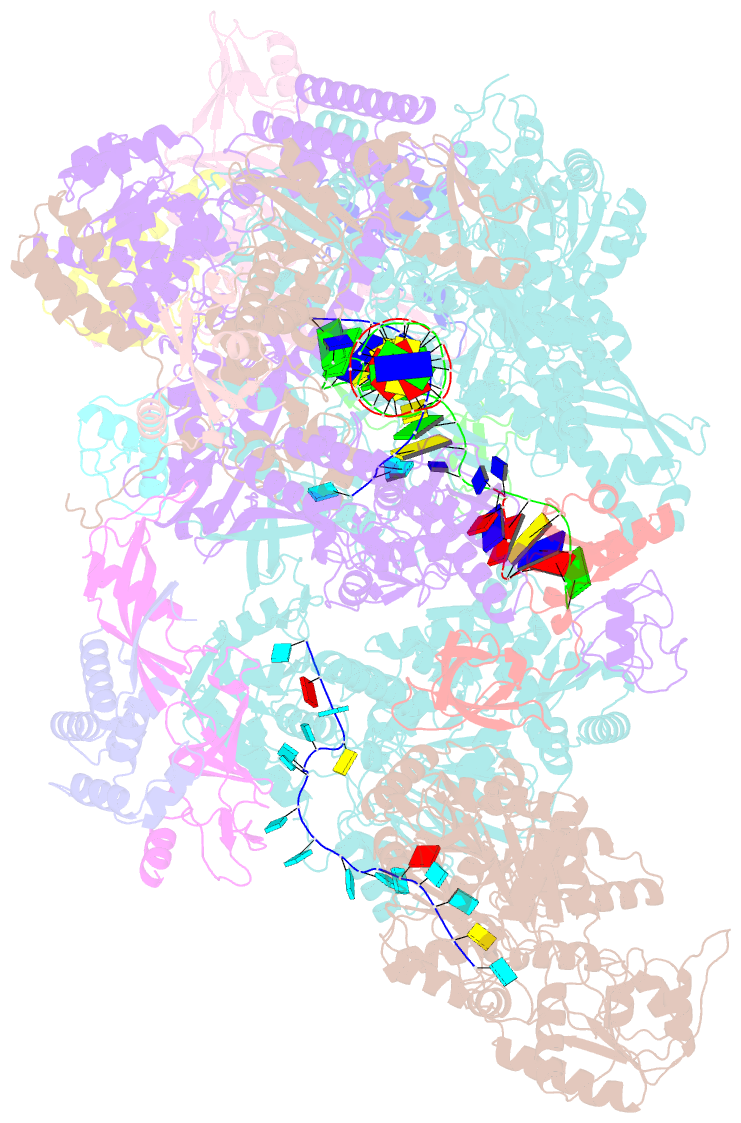
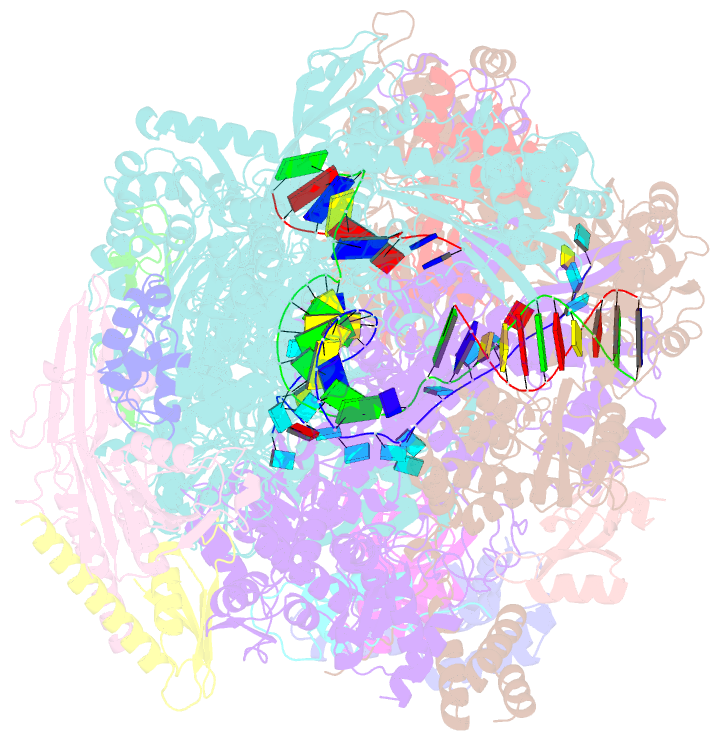
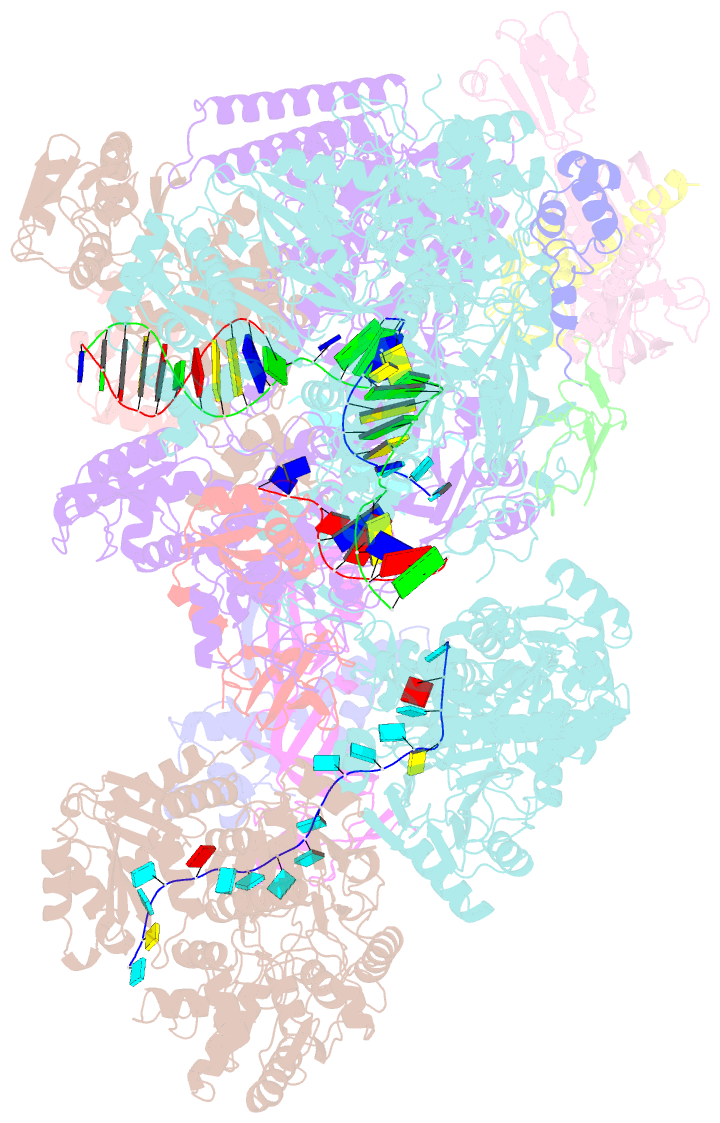
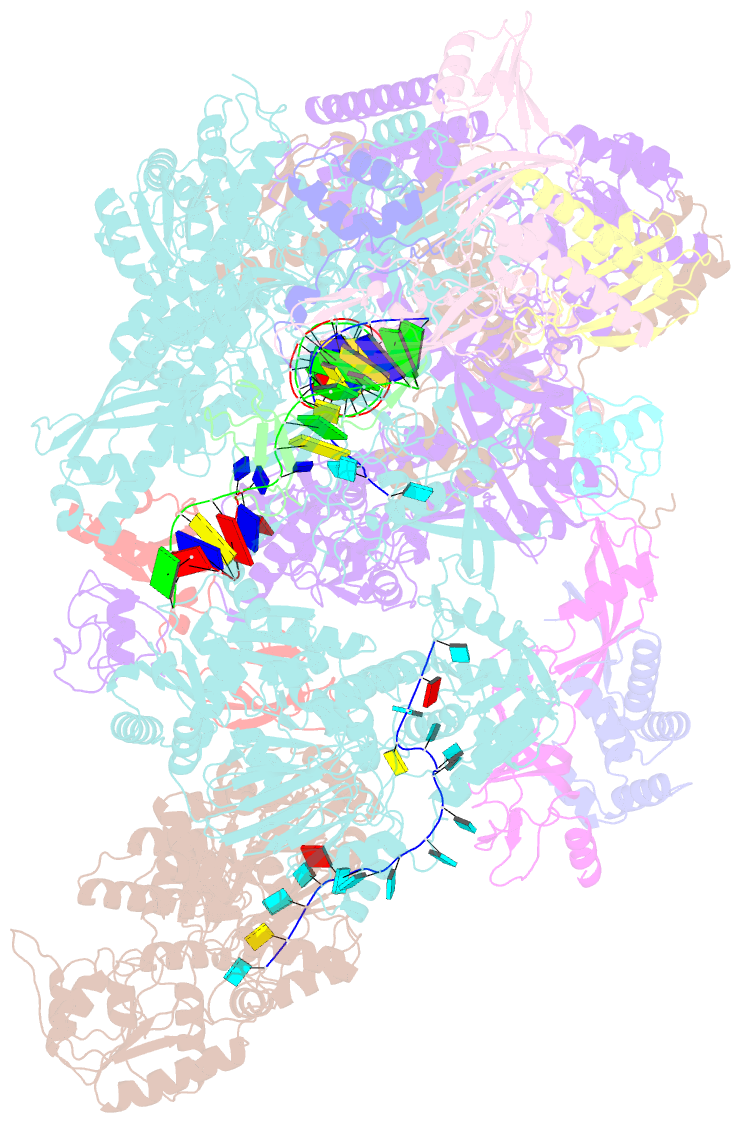
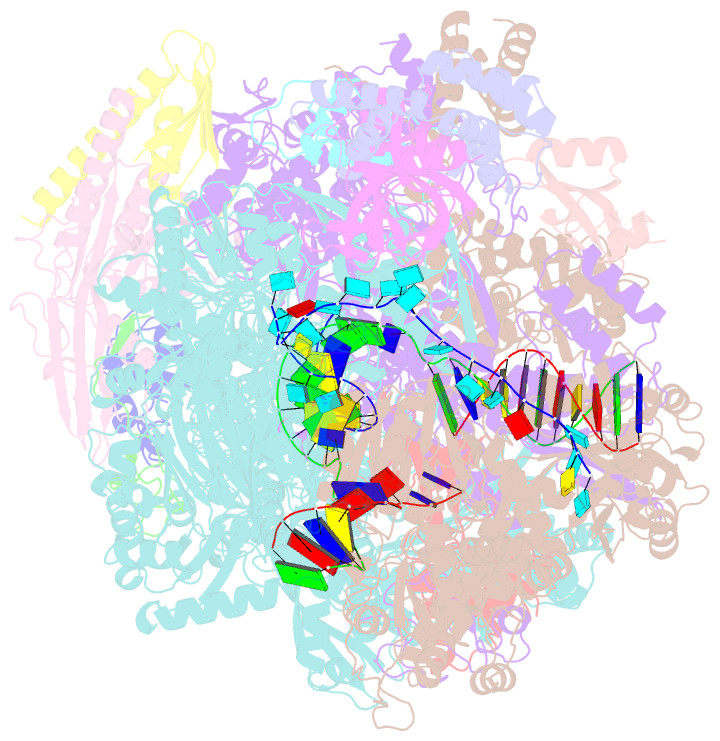
- cryo-EM structure of thermococcus kodakarensis ftta-dependent transcription pre-termination complex containing 44 nt RNA
- Reference
- You L, Wang C, Molodtsov V, Kuznedelov K, Miao X, Wenck BR, Ulisse P, Sanders TJ, Marshall CJ, Firlar E, Kaelber JT, Santangelo TJ, Ebright RH (2024): "Structural basis of archaeal FttA-dependent transcription termination." Nature, 635, 229-236. doi: 10.1038/s41586-024-07979-9.
- Abstract
- The ribonuclease FttA (also known as aCPSF and aCPSF1) mediates factor-dependent transcription termination in archaea1-3. Here we report the structure of a Thermococcus kodakarensis transcription pre-termination complex comprising FttA, Spt4, Spt5 and a transcription elongation complex (TEC). The structure shows that FttA interacts with the TEC in a manner that enables RNA to proceed directly from the TEC RNA-exit channel to the FttA catalytic centre and that enables endonucleolytic cleavage of RNA by FttA, followed by 5'→3' exonucleolytic cleavage of RNA by FttA and concomitant 5'→3' translocation of FttA on RNA, to apply mechanical force to the TEC and trigger termination. The structure further reveals that Spt5 bridges FttA and the TEC, explaining how Spt5 stimulates FttA-dependent termination. The results reveal functional analogy between bacterial and archaeal factor-dependent termination, functional homology between archaeal and eukaryotic factor-dependent termination, and fundamental mechanistic similarities in factor-dependent termination in bacteria, archaea, and eukaryotes.