Summary information and primary citation
- PDB-id
- 9c5q; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- cryo-EM (3.1 Å)
- Summary
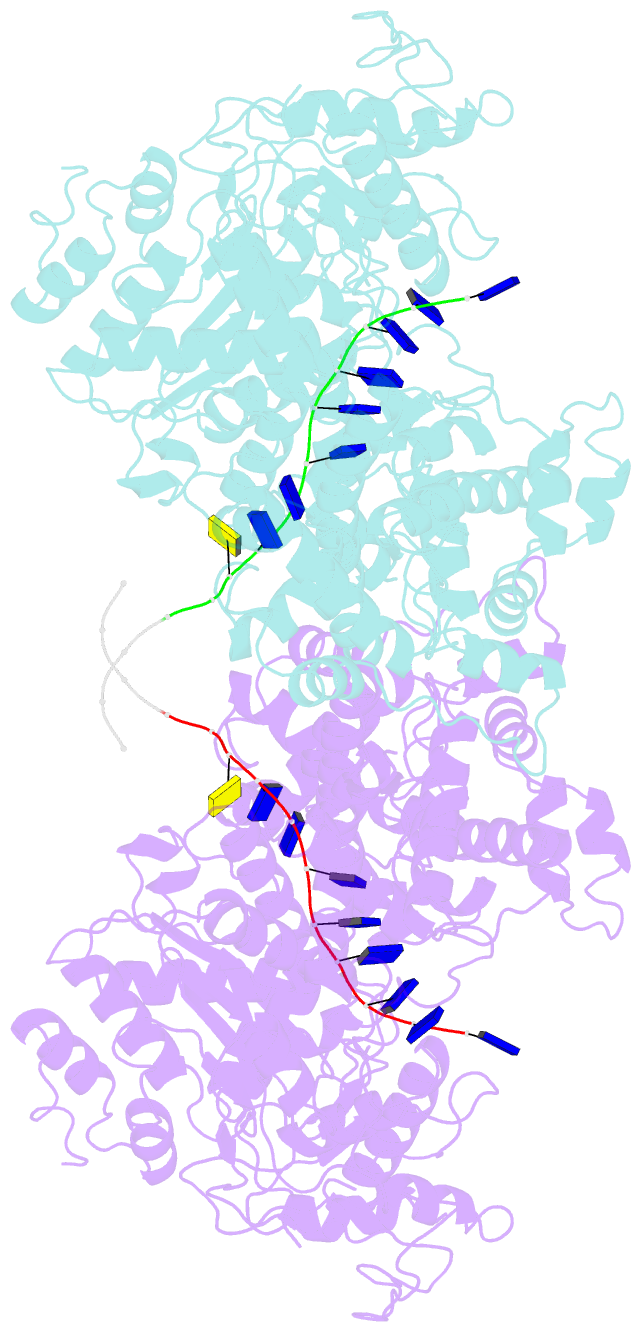
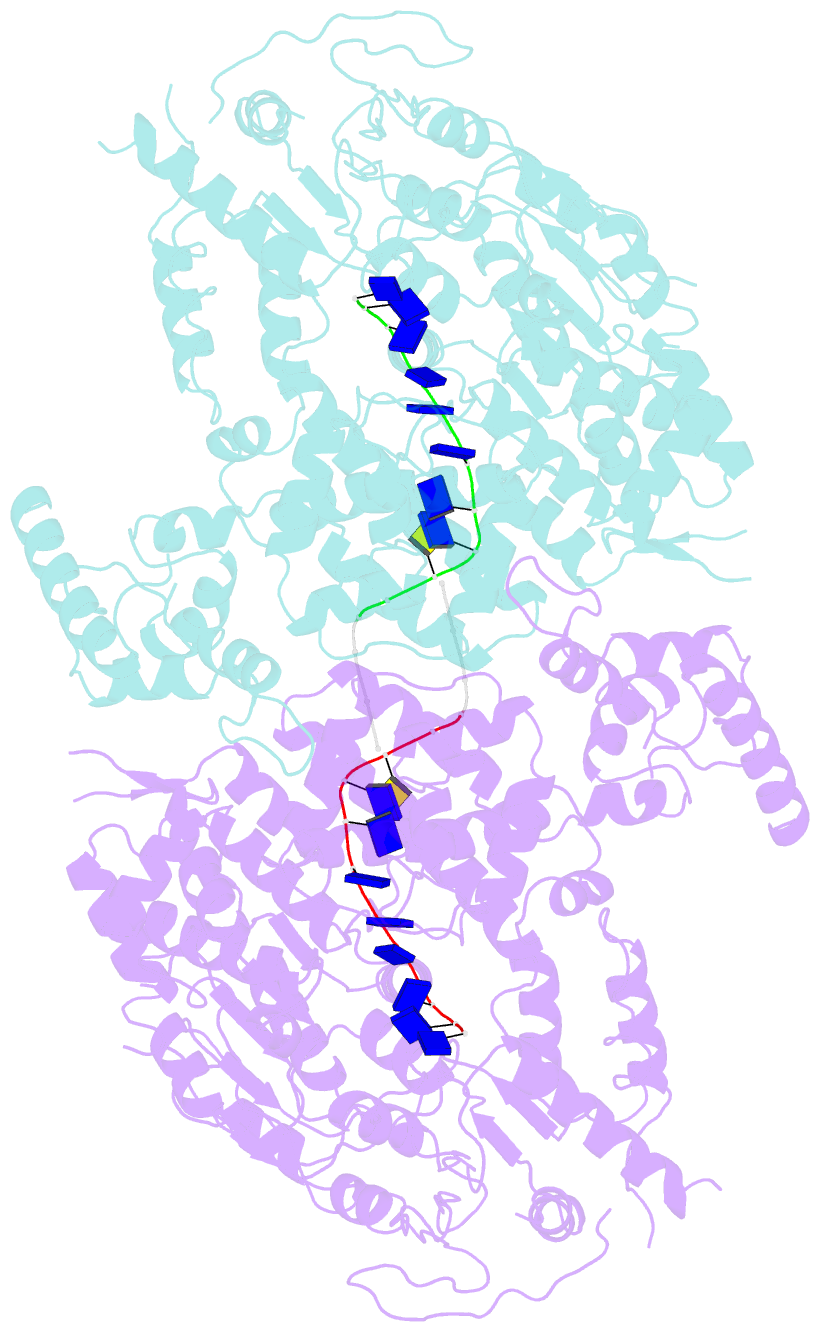
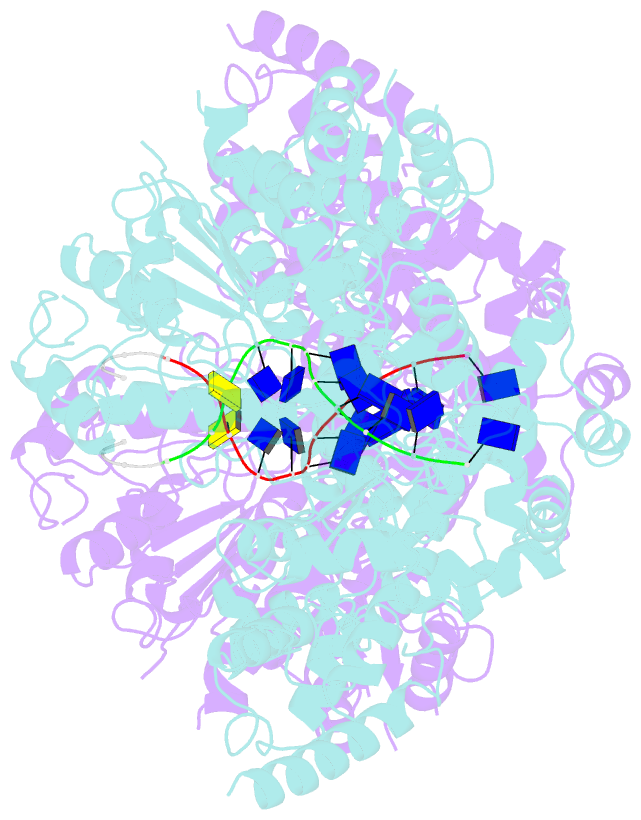
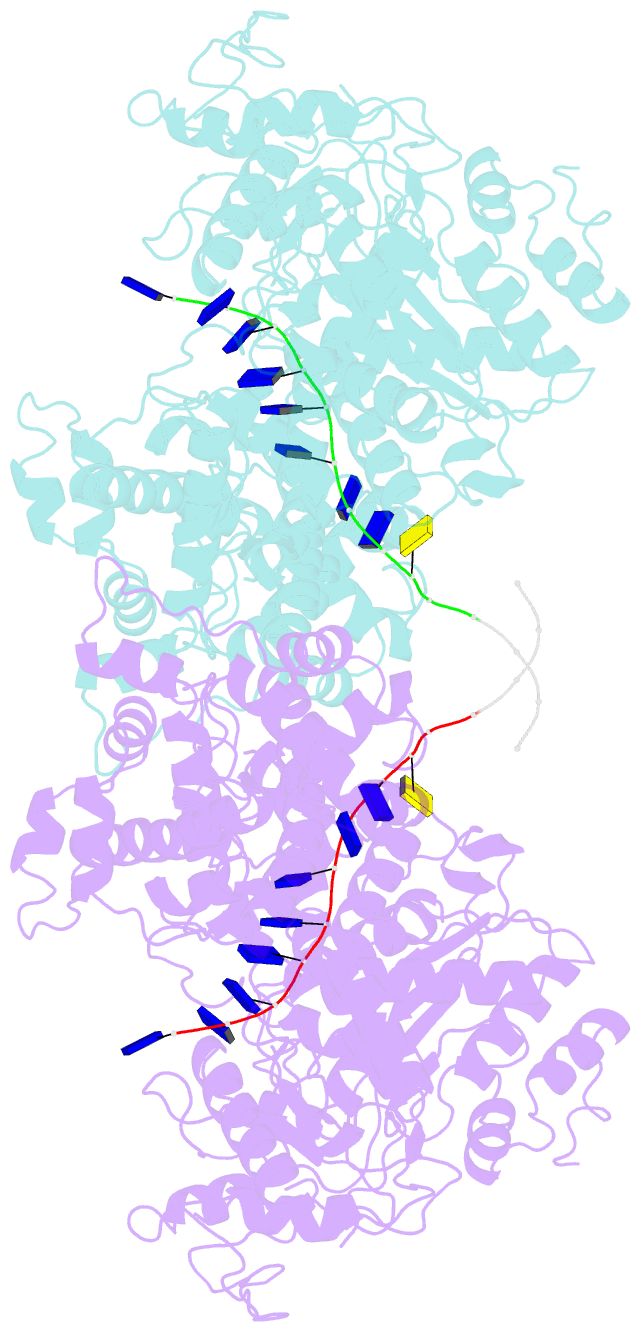
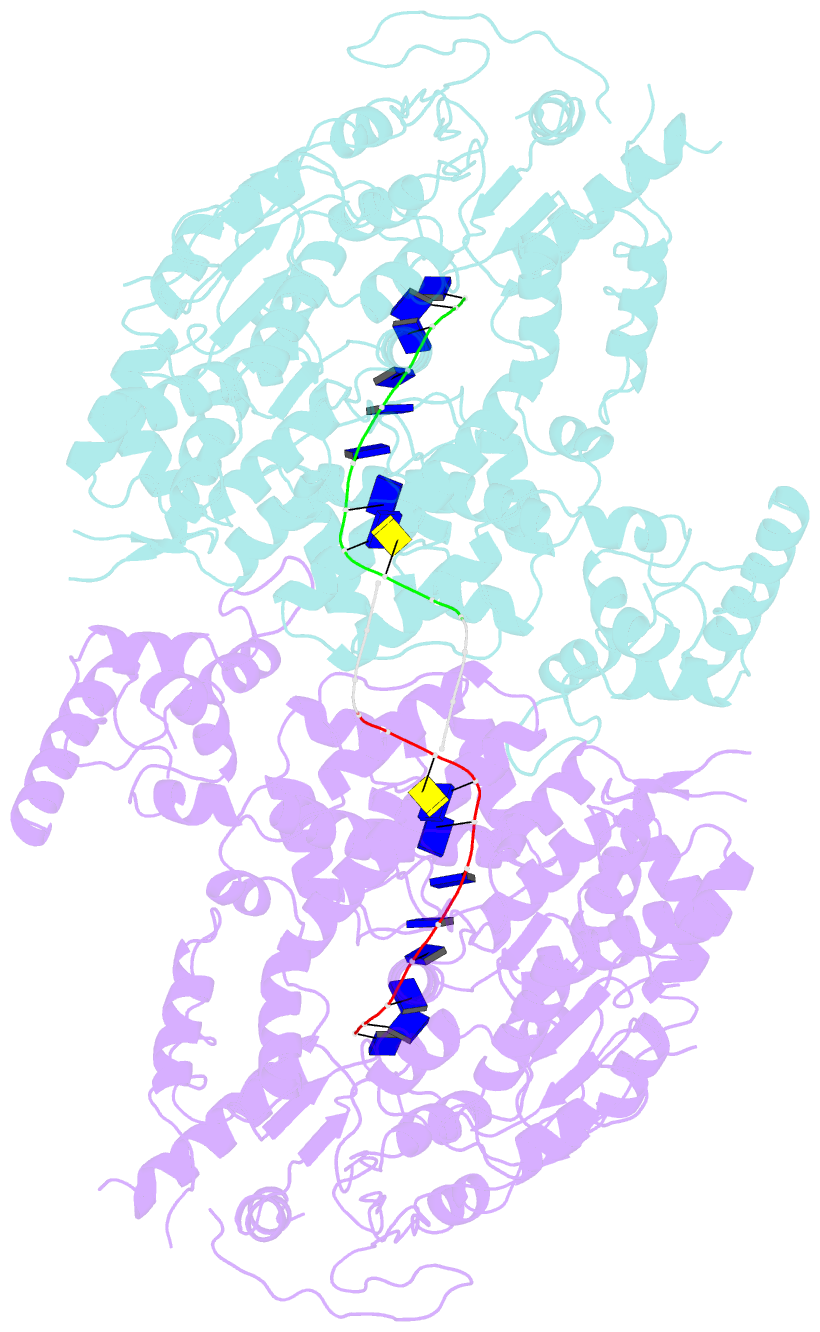
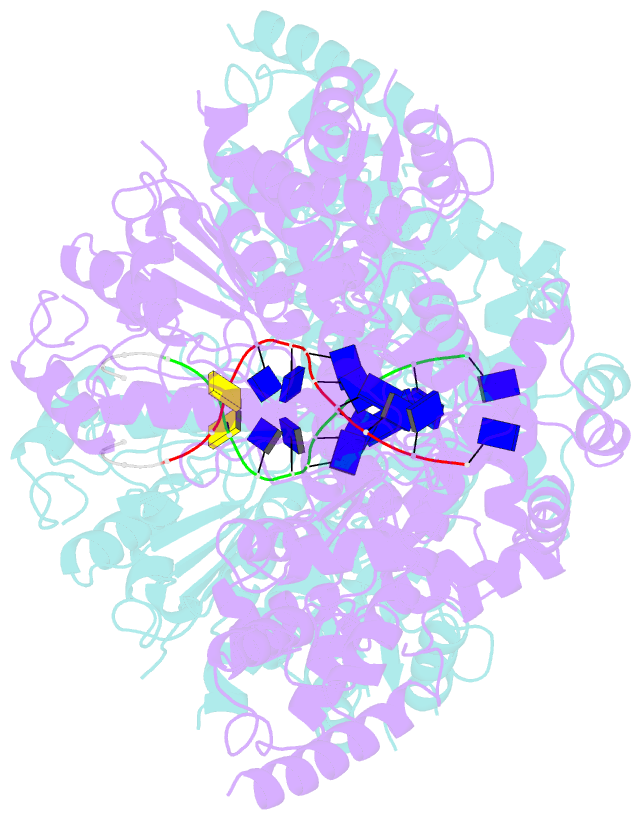
- Human DNA polymerase theta helicase domain in microhomology annealed state 2, dimer form
- Reference
- Ito F, Li Z, Minakhin L, Khant HA, Pomerantz RT, Chen XS (2024): "Structural Basis for Pol theta-Helicase DNA Binding and Microhomology-Mediated End-Joining." Biorxiv. doi: 10.1101/2024.06.07.597860.
- Abstract
- DNA double-strand breaks (DSBs) present a critical threat to genomic integrity, often precipitating genomic instability and oncogenesis. Repair of DSBs predominantly occurs through homologous recombination (HR) and non-homologous end joining (NHEJ). In HR-deficient cells, DNA polymerase theta (Polθ) becomes critical for DSB repair via microhomology-mediated end joining (MMEJ), also termed theta-mediated end joining (TMEJ). Thus, Polθ is synthetically lethal with BRCA1/2 and other HR factors, underscoring its potential as a therapeutic target in HR-deficient cancers. However, the molecular mechanisms governing Polθ-mediated MMEJ remain poorly understood. Here we present a series of cryo-electron microscopy structures of the Polθ helicase domain (Polθ-hel) in complex with DNA containing 3'-overhang. The structures reveal the sequential conformations adopted by Polθ-hel during the critical phases of DNA binding, microhomology searching, and microhomology annealing. The stepwise conformational changes within the Polθ-hel subdomains and its functional dimeric state are pivotal for aligning the 3'-overhangs, facilitating the microhomology search and subsequent annealing necessary for DSB repair via MMEJ. Our findings illustrate the essential molecular switches within Polθ-hel that orchestrate the MMEJ process in DSB repair, laying the groundwork for the development of targeted therapies against the Polθ-hel.