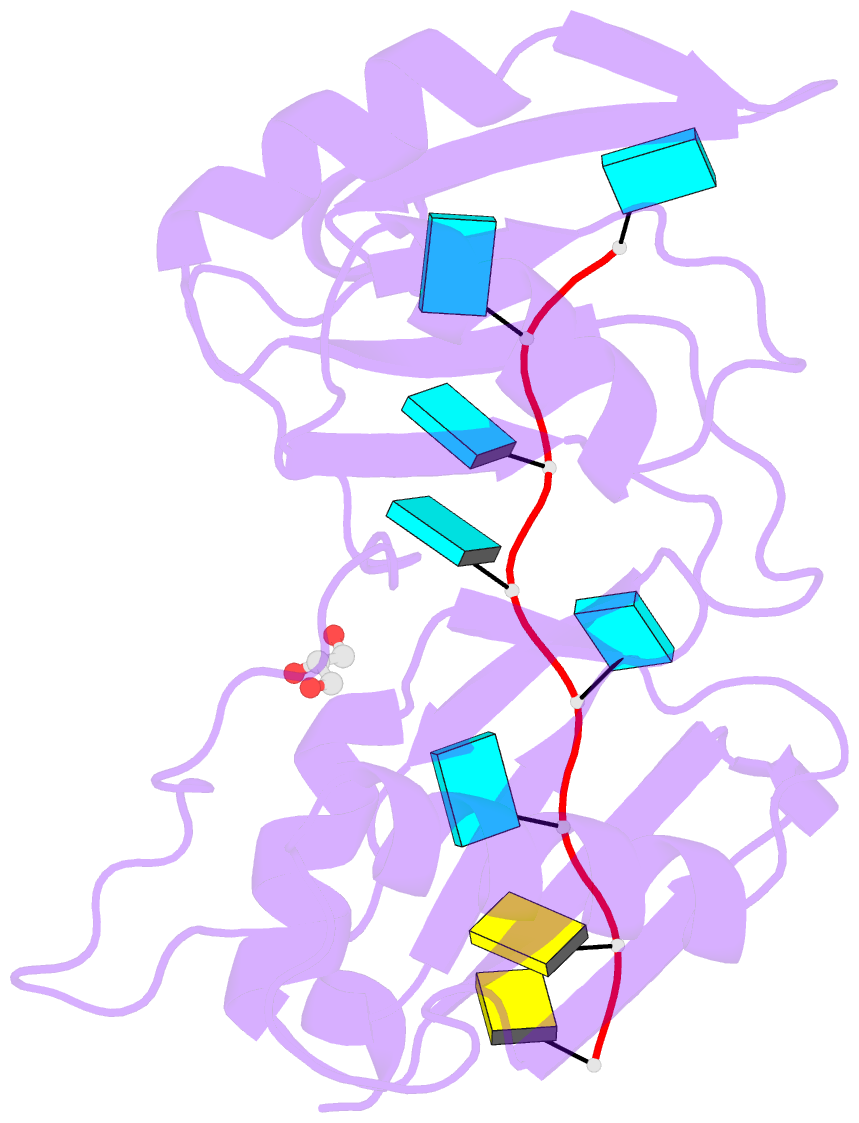
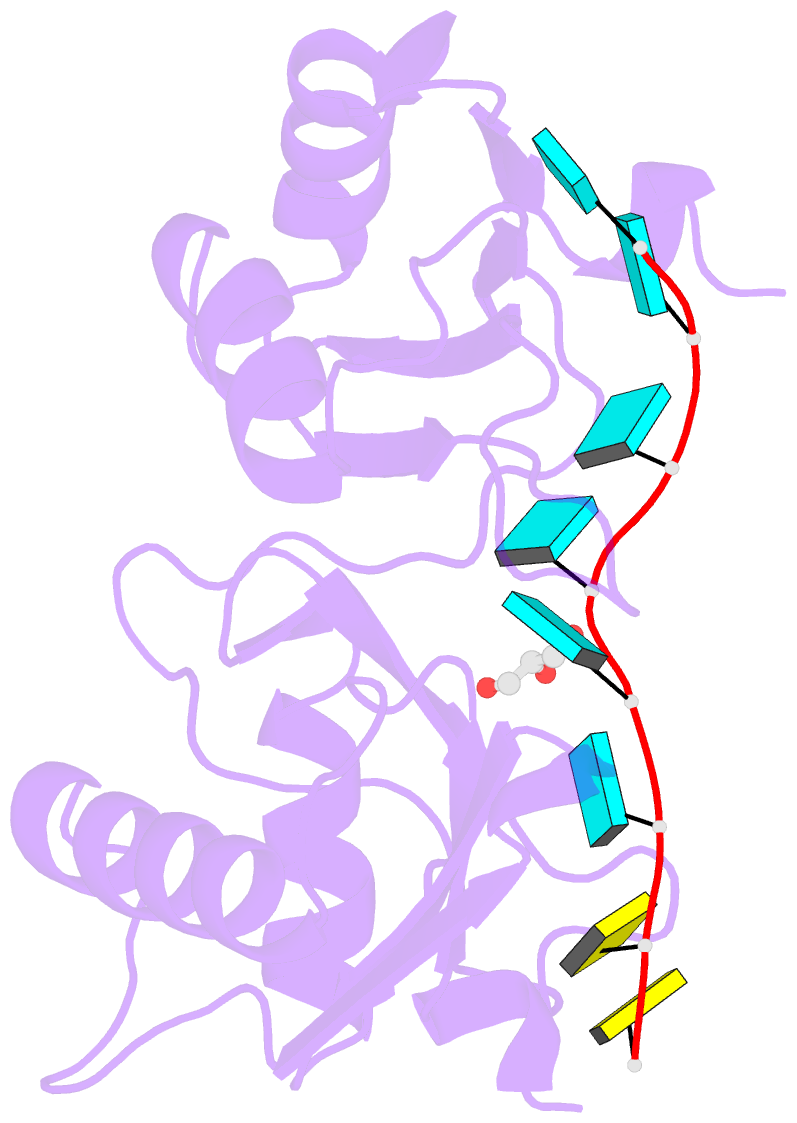
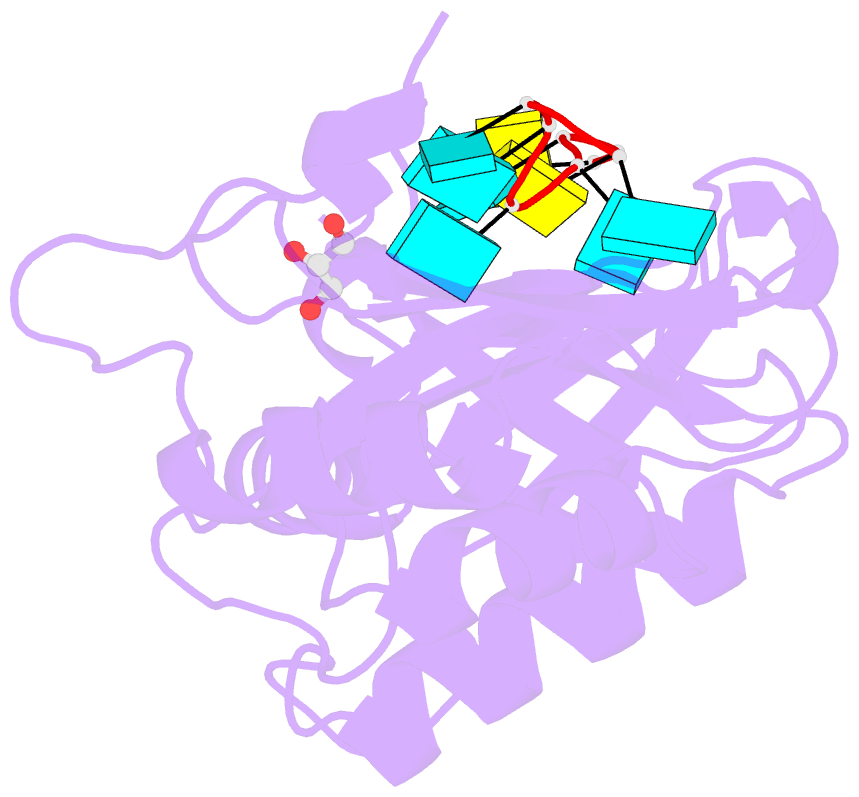
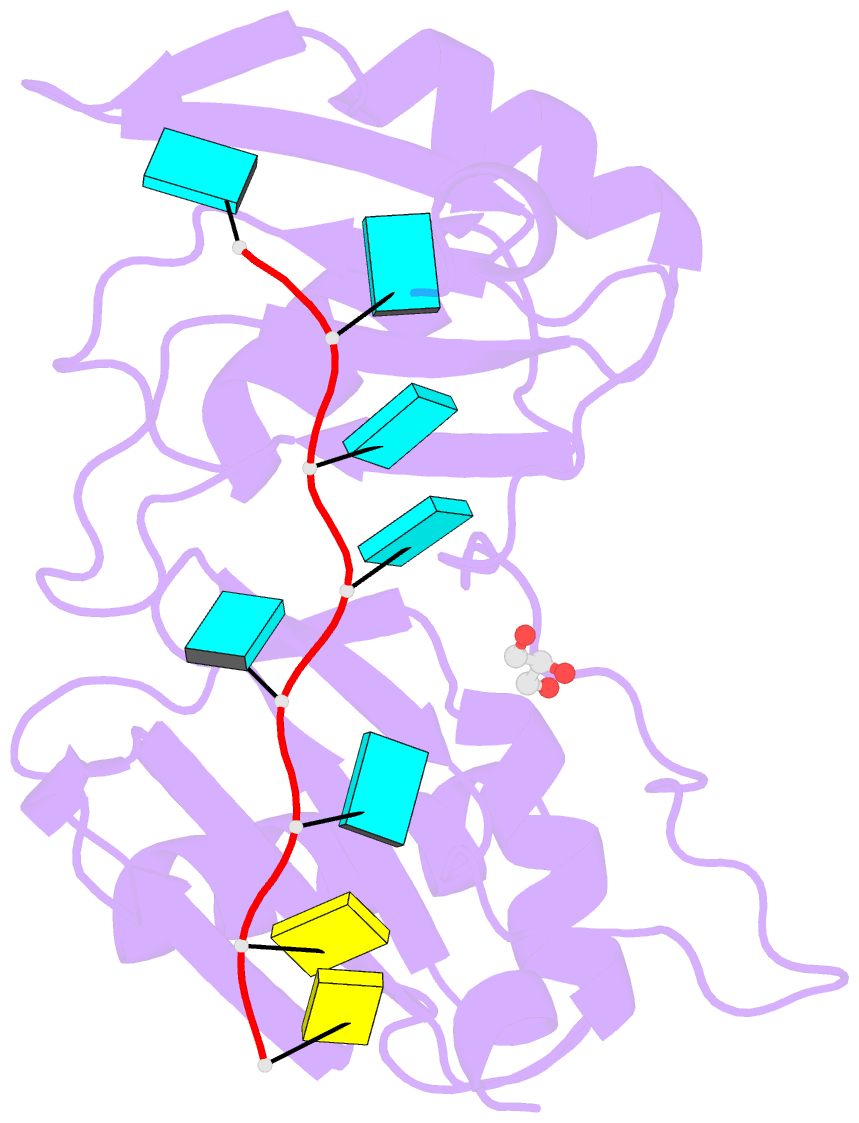
Summary information and primary citation
- PDB-id
- 9c7a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (1.4 Å)
- Summary
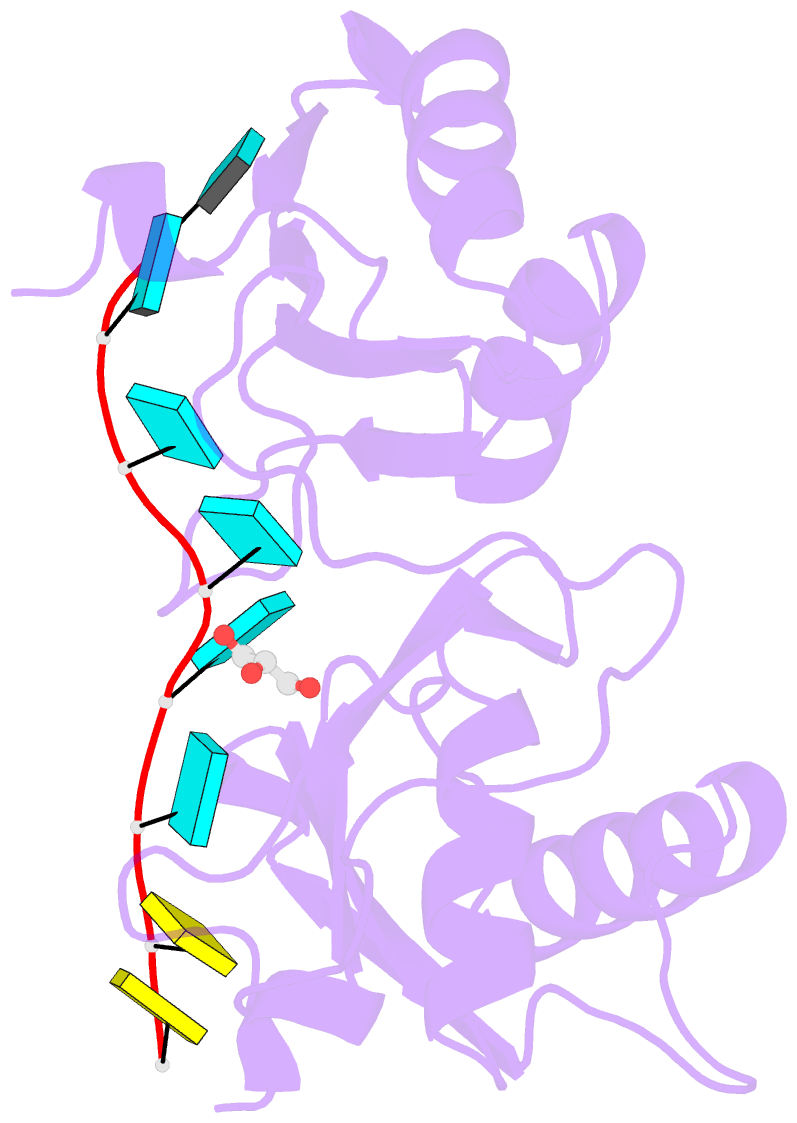
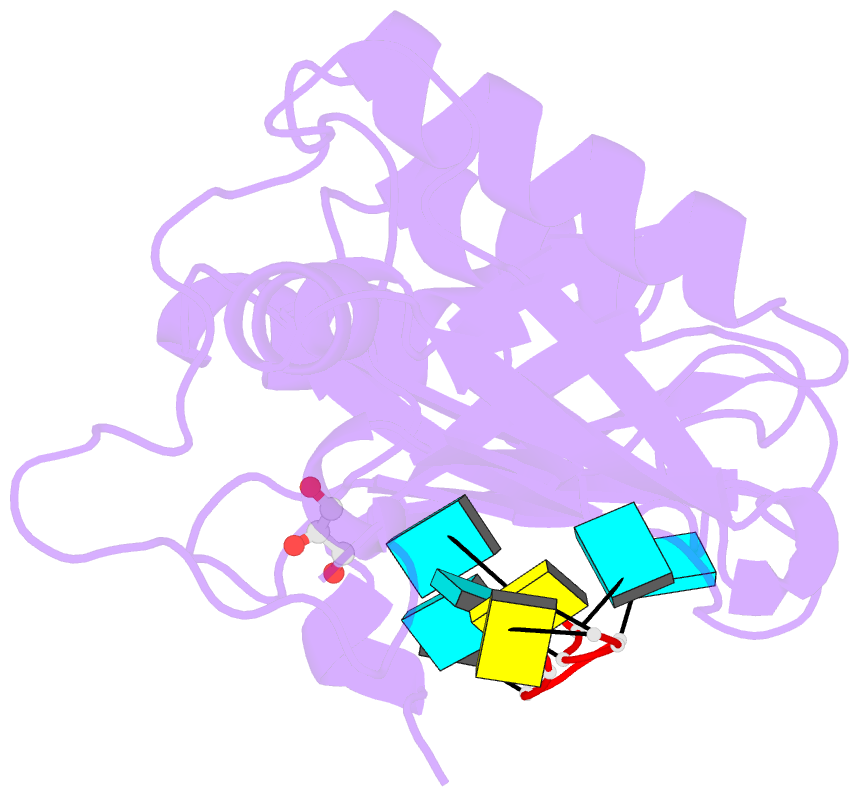
- Crystal structure of r149w neurodevelopmental disease-associated u2af2 variant
- Reference
- Maji D, Jenkins JL, Boutz PL, Kielkopf CL (2024): "Recurrent Neurodevelopmentally Associated Variants of the Pre-mRNA Splicing Factor U2AF2 Alter RNA Binding Affinities and Interactions." Biochemistry. doi: 10.1021/acs.biochem.4c00344.
- Abstract
- De novo mutations affecting the pre-mRNA splicing factor U2AF2 are associated with developmental delays and intellectual disabilities, yet the molecular basis is unknown. Here, we demonstrated by fluorescence anisotropy RNA binding assays that recurrent missense mutants (Arg149Trp, Arg150His, or Arg150Cys) decreased the binding affinity of U2AF2 for a consensus splice site RNA. Crystal structures at 1.4 Å resolutions showed that Arg149Trp or Arg150His disrupted hydrogen bonds between U2AF2 and the terminal nucleotides of the RNA site. Reanalysis of publicly available RNaseq data confirmed that U2AF2 depletion altered splicing of transcripts encoding RNA binding proteins (RBPs). These results confirmed that the impaired RNA interactions of Arg149Trp and Arg150His U2AF2 variants could contribute to dysregulating an RBP-governed neurodevelopmental program of alternative splicing.