Summary information and primary citation
- PDB-id
- 9cdb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- antiviral protein-RNA
- Method
- cryo-EM (3.6 Å)
- Summary
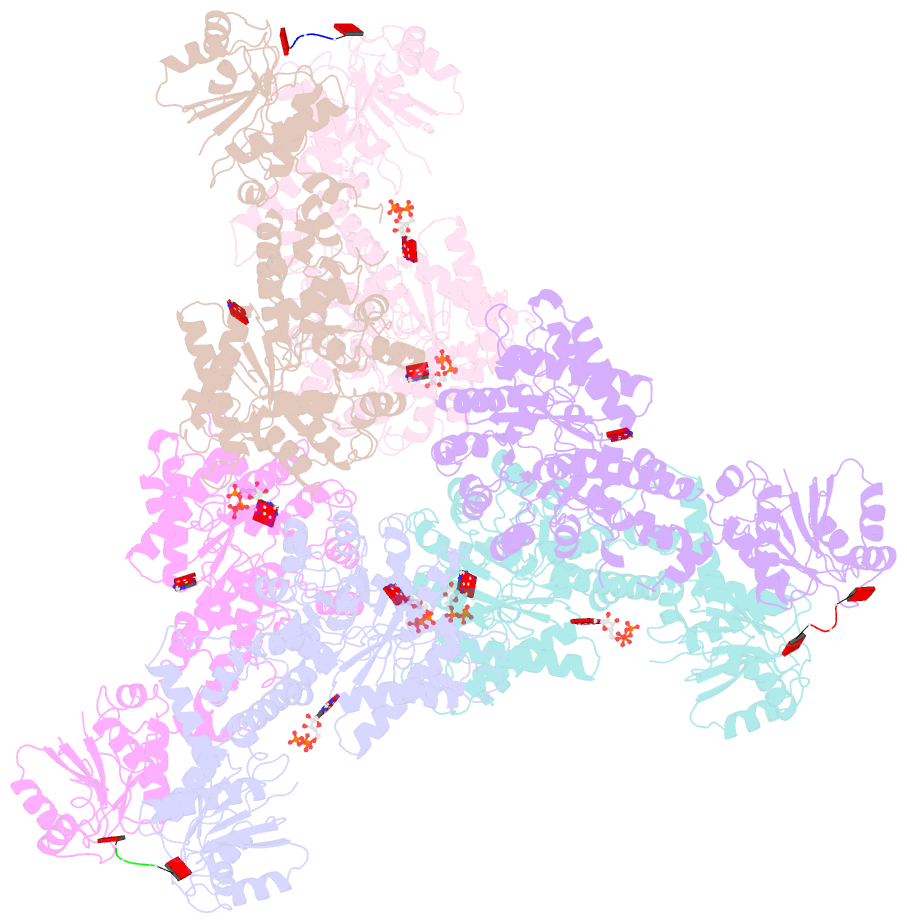
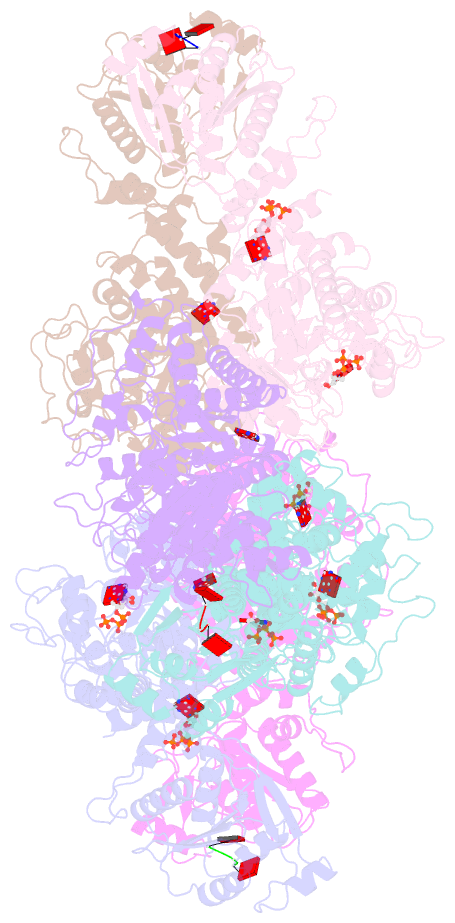
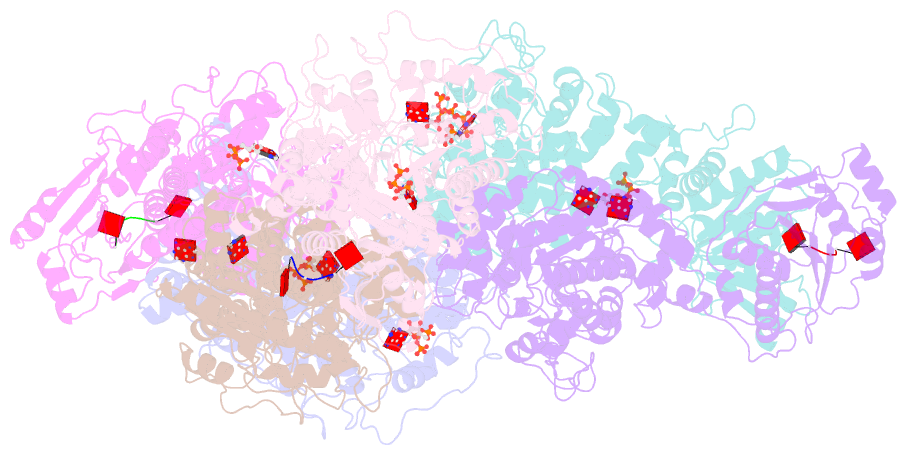
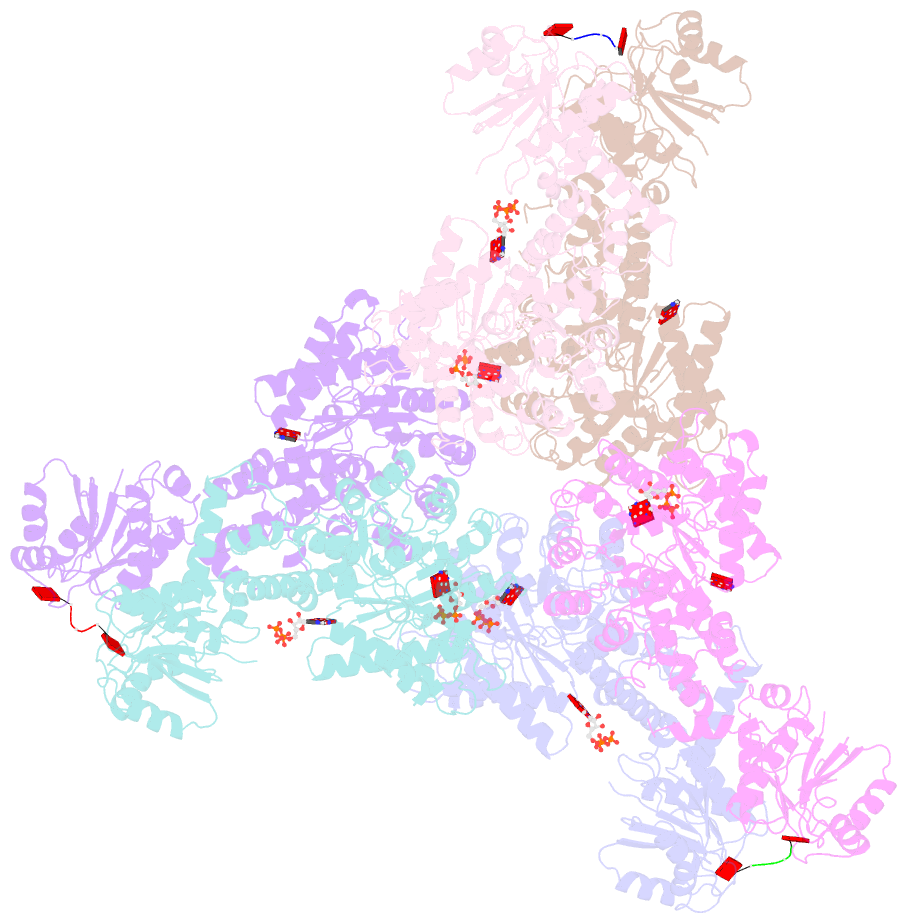
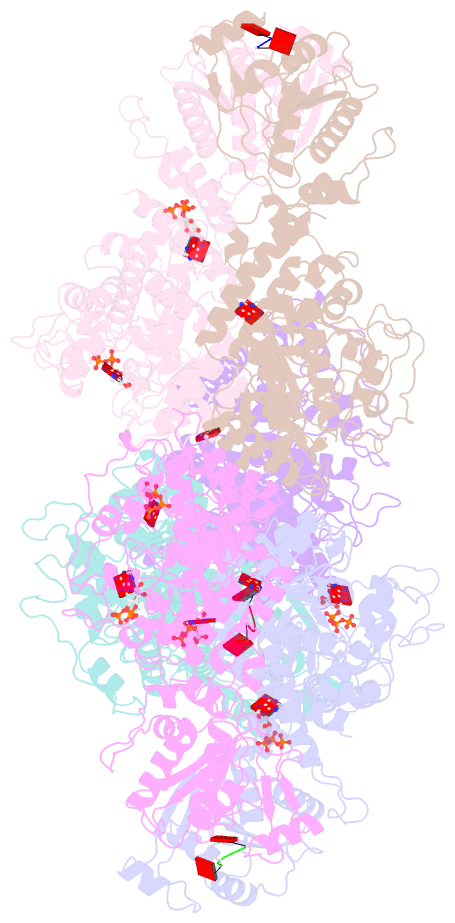
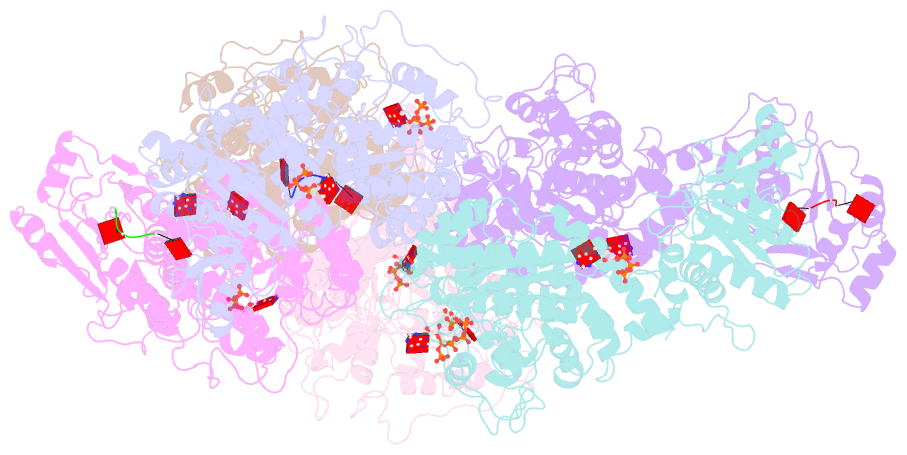
- Cryoem structure of crispr associated effector, carf-adenosine deaminase 1, cad1, in ca6 (partial density) bound form with atp (partial density).
- Reference
- Baca CF, Majumder P, Hickling JH, Ye L, Teplova M, Brady SF, Patel DJ, Marraffini LA (2024): "The CRISPR-associated adenosine deaminase Cad1 converts ATP to ITP to provide antiviral immunity." Cell. doi: 10.1016/j.cell.2024.10.002.
- Abstract
- Type III CRISPR systems provide immunity against genetic invaders through the production of cyclic oligo-adenylate (cAn) molecules that activate effector proteins that contain CRISPR-associated Rossman fold (CARF) domains. Here, we characterized the function and structure of an effector in which the CARF domain is fused to an adenosine deaminase domain, CRISPR-associated adenosine deaminase 1 (Cad1). We show that upon binding of cA4 or cA6 to its CARF domain, Cad1 converts ATP to ITP, both in vivo and in vitro. Cryoelectron microscopy (cryo-EM) structural studies on full-length Cad1 reveal an hexameric assembly composed of a trimer of dimers, with bound ATP at inter-domain sites required for activity and ATP/ITP within deaminase active sites. Upon synthesis of cAn during phage infection, Cad1 activation leads to a growth arrest of the host that prevents viral propagation. Our findings reveal that CRISPR-Cas systems employ a wide range of molecular mechanisms beyond nucleic acid degradation to provide adaptive immunity in prokaryotes.