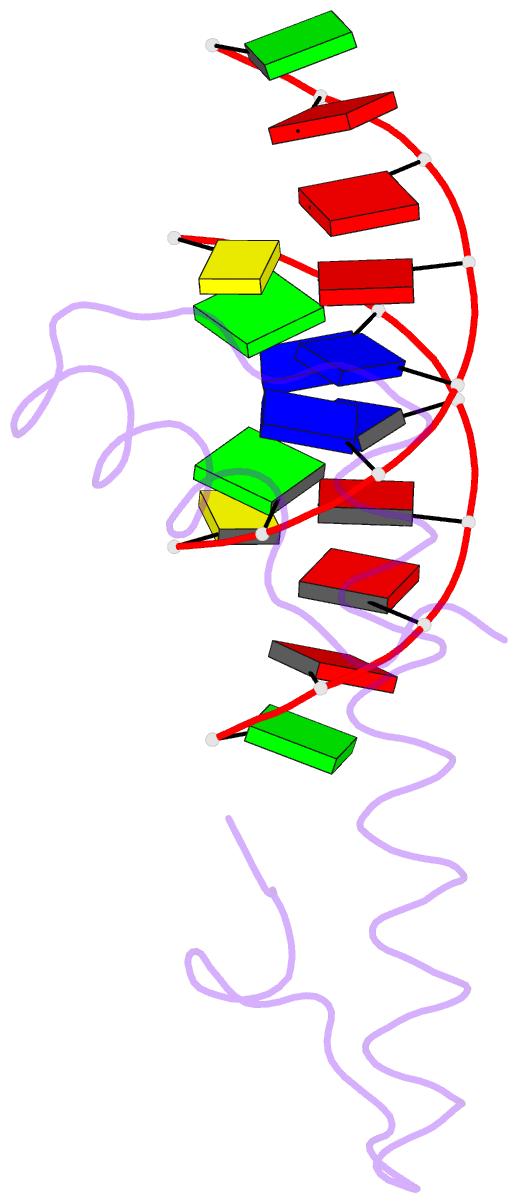
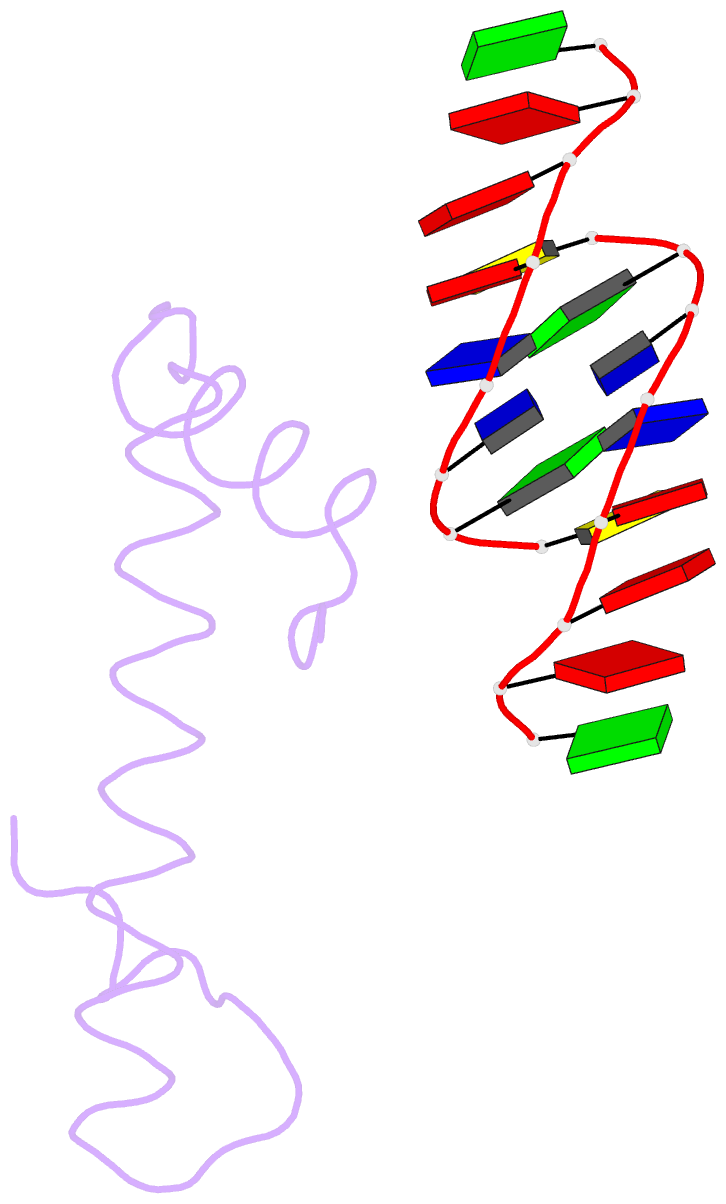
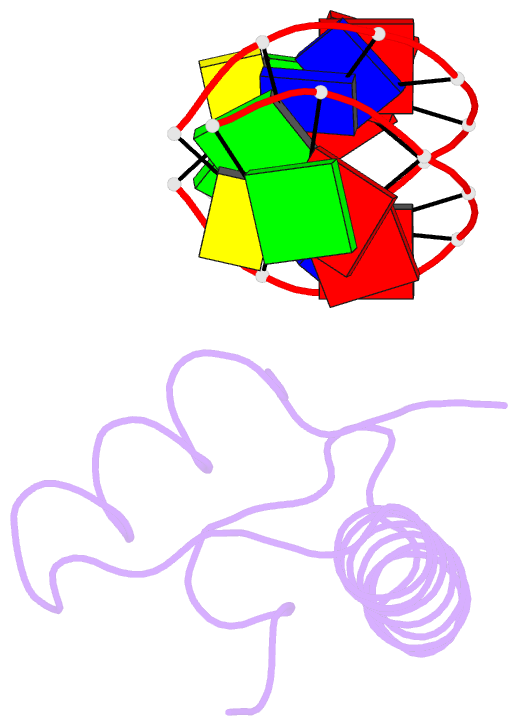
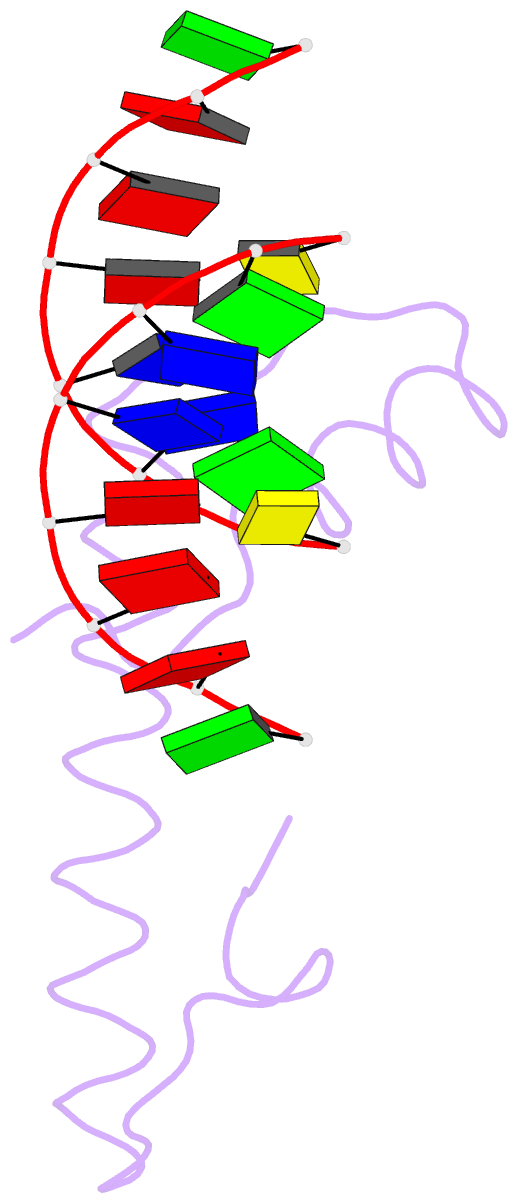
Summary information and primary citation
- PDB-id
- 9f0e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein
- Method
- X-ray (1.85 Å)
- Summary
- Bacterial histone protein hbb from bdellovibrio bacteriovorus bound to DNA
- Reference
- Hu Y, Schwab S, Deiss S, Escudeiro P, van Heesch T, Joiner JD, Vreede J, Hartmann MD, Lupas AN, Alvarez BH, Alva V, Dame RT (2024): "Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending." Nucleic Acids Res., 52, 8193-8204. doi: 10.1093/nar/gkae485.
- Abstract
- Histones, traditionally known for organizing and regulating DNA in eukaryotes and archaea, have recently been discovered in bacteria, opening up a new frontier in our understanding of genome organization across the domains of life. Our study investigates the largely unexplored DNA-binding properties of bacterial histones, focusing on HBb in Bdellovibrio bacteriovorus. We reveal that HBb is essential for bacterial survival and exhibits DNA-binding properties similar to archaeal and eukaryotic histones. However, unlike eukaryotic and archaeal histones, which wrap DNA, HBb bends DNA without sequence specificity. This work not only broadens our understanding of DNA organization across different life forms but also suggests that bacterial histones may have diverse roles in genome organization.